- Record: found
- Abstract: found
- Article: found
A Patient with Berardinelli-Seip Syndrome, Novel AGPAT2 Splicesite Mutation and Concomitant Development of Non-diabetic Polyneuropathy
Read this article at
Abstract
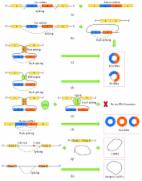
Primary polyneuropathy in the context of Seip-Berardinelli type 1 seipinopathy, or congenital generalized lipodystrophy type 1 (CGL1) has not been previously reported. We report the case history of a 27 year old female CGL1 patient presenting with an unusual additional development of non-diabetic peripheral neuropathy and learning disabilities in early adolescence. Whole exome sequencing (WES) of the patient genome identified a novel variant, homozygous for a 52 bp intronic deletion in the AGPAT2 locus, coding for 1-acylglycerol-3-phosphate O-acyltransferase 2, which is uniquely associated with CGL1 seipinopathies, with no molecular evidence for dual diagnosis. Functional studies using RNA isolated from patient peripheral blood leucocytes showed abnormal RNA splicing resulting in the loss of 25 amino acids from the patient AGPAT2 protein coding sequence. Stability and transcription levels for the misspliced AGPAT2 mRNA in our patient nonetheless remained normal. Any AGPAT2 protein produced in our patient is therefore likely to be dysfunctional. However, formal linkage of this deletion to the neuropathy observed remains to be shown. The classical clinical presentation of a patient with AGPAT2-associated lipodystrophy shows normal cognition and no development of polyneuropathy. Cognitive disabilities and polyneuropathy are features associated exclusively with clinical CGL type 2 arising from seipin ( BSCL2) gene mutations. This case study suggests that in some genetic contexts, AGPAT2 mutations can also produce phenotypes with primary polyneuropathy.
Related collections
Most cited references12
- Record: found
- Abstract: found
- Article: not found
Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation.
- Record: found
- Abstract: found
- Article: not found
Using XHMM Software to Detect Copy Number Variation in Whole-Exome Sequencing Data.
- Record: found
- Abstract: found
- Article: found