- Record: found
- Abstract: found
- Article: found
Hen raising helps chicks establish gut microbiota in their early life and improve microbiota stability after H9N2 challenge
Read this article at
Abstract
Background
Early gut microbial colonization is important for postnatal growth and immune development of the chicken. However, at present, commercial chickens are hatched and raised without adult hens, thus are cut off from the microbiota transfer between hens and chicks. In this study, we compared the gut microbiota composition between hen-reared and separately reared chicks, and its impact on the resistance to H9N2 avian influenza virus, with the motive of investigating the impact of this cutoff in microbiota transfer.
Results
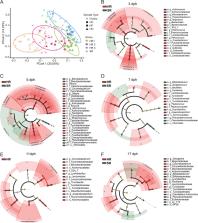
We used the 16SrRNA sequencing method to assess the composition of the gut microbiota in chicks represented by three hen-reared groups and one separately reared group. We found that the diversity of gut microbes in the chicks from the three hen-reared groups was more abundant than in the separately reared group, both at the phylum and genus levels. Our findings highlight the importance of early parental care in influencing the establishment of gut microbiota in the early life of chicks. SourceTracker analysis showed that the feather and cloaca microbiota of hens are the main sources of gut microbiota of chicks. After H9N2 exposure, the viral infection lasted longer in the separately reared chicks, with the viral titers in their oropharyngeal swabs being higher compared to the hen-reared chicks at day 5 post-infection. Interestingly, our results revealed that the gut microbiota of the hen-reared chicks was more stable after H9N2 infection in comparison to that of the separately reared chicks.
Conclusions
Microbiota transfer between the hens and their chicks promotes the establishment of a balanced and diverse microbiota in the early life of the chicks and improves microbiota stability after H9N2 challenge. These findings advance our understanding of the protective role of gut microbiota in the early life of chicks and should be instrumental in improving chick rearing in the commercial poultry industry.
Related collections
Most cited references42

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: not found
- Article: not found
Cutadapt removes adapter sequences from high-throughput sequencing reads
- Record: found
- Abstract: found
- Article: not found