- Record: found
- Abstract: found
- Article: found
Identification of a six-gene signature predicting overall survival for hepatocellular carcinoma
Read this article at
Abstract
Background
Hepatocellular carcinoma (HCC) remains a major challenge for public health worldwide. Considering the great heterogeneity of HCC, more accurate prognostic models are urgently needed. To identify a robust prognostic gene signature, we conduct this study.
Materials and methods
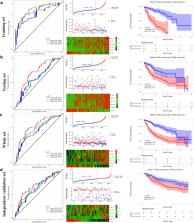
Level 3 mRNA expression profiles and clinicopathological data were obtained in The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC). GSE14520 dataset from the gene expression omnibus (GEO) database was downloaded to further validate the results in TCGA. Differentially expressed mRNAs between HCC and normal tissue were investigated. Univariate Cox regression analysis and lasso Cox regression model were performed to identify and construct the prognostic gene signature. Time-dependent receiver operating characteristic (ROC), Kaplan–Meier curve, multivariate Cox regression analysis, nomogram, and decision curve analysis (DCA) were used to assess the prognostic capacity of the six-gene signature. The prognostic value of the gene signature was further validated in independent GSE14520 cohort. Gene Set Enrichment Analyses (GSEA) was performed to further understand the underlying molecular mechanisms. The performance of the prognostic signature in differentiating between normal liver tissues and HCC were also investigated.
Results
A novel six-gene signature (including CSE1L, CSTB, MTHFR, DAGLA, MMP10, and GYS2) was established for HCC prognosis prediction. The ROC curve showed good performance in survival prediction in both the TCGA HCC cohort and the GSE14520 validation cohort. The six-gene signature could stratify patients into a high- and low-risk group which had significantly different survival. Cox regression analysis showed that the six-gene signature could independently predict OS. Nomogram including the six-gene signature was established and shown some clinical net benefit. Furthermore, GSEA revealed several significantly enriched oncological signatures and various metabolic process, which might help explain the underlying molecular mechanisms. Besides, the prognostic signature showed a strong ability for differentiating HCC from normal tissues.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.
- Record: found
- Abstract: found
- Article: not found
Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain
- Record: found
- Abstract: found
- Article: not found