- Record: found
- Abstract: found
- Article: found
Gene editing and CRISPR in the clinic: current and future perspectives
Read this article at
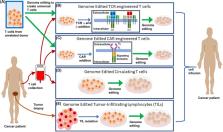
Abstract
Genome editing technologies, particularly those based on zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR (clustered regularly interspaced short palindromic repeat DNA sequences)/Cas9 are rapidly progressing into clinical trials. Most clinical use of CRISPR to date has focused on ex vivo gene editing of cells followed by their re-introduction back into the patient. The ex vivo editing approach is highly effective for many disease states, including cancers and sickle cell disease, but ideally genome editing would also be applied to diseases which require cell modification in vivo. However, in vivo use of CRISPR technologies can be confounded by problems such as off-target editing, inefficient or off-target delivery, and stimulation of counterproductive immune responses. Current research addressing these issues may provide new opportunities for use of CRISPR in the clinical space. In this review, we examine the current status and scientific basis of clinical trials featuring ZFNs, TALENs, and CRISPR-based genome editing, the known limitations of CRISPR use in humans, and the rapidly developing CRISPR engineering space that should lay the groundwork for further translation to clinical application.
Related collections
Most cited references171
- Record: found
- Abstract: found
- Article: not found
Epigenome editing by a CRISPR/Cas9-based acetyltransferase activates genes from promoters and enhancers
- Record: found
- Abstract: found
- Article: found
CRISPR–Cas9 Structures and Mechanisms
- Record: found
- Abstract: found
- Article: not found