- Record: found
- Abstract: found
- Article: found
Targeting autophagy-related protein kinases for potential therapeutic purpose

Read this article at
Abstract
Autophagy, defined as a scavenging process of protein aggregates and damaged organelles mediated by lysosomes, plays a significant role in the quality control of macromolecules and organelles. Since protein kinases are integral to the autophagy process, it is critically important to understand the role of kinases in autophagic regulation. At present, intervention of autophagic processes by small-molecule modulators targeting specific kinases has becoming a reasonable and prevalent strategy for treating several varieties of human disease, especially cancer. In this review, we describe the role of some autophagy-related kinase targets and kinase-mediated phosphorylation mechanisms in autophagy regulation. We also summarize the small-molecule kinase inhibitors/activators of these targets, highlighting the opportunities of these new therapeutic agents.
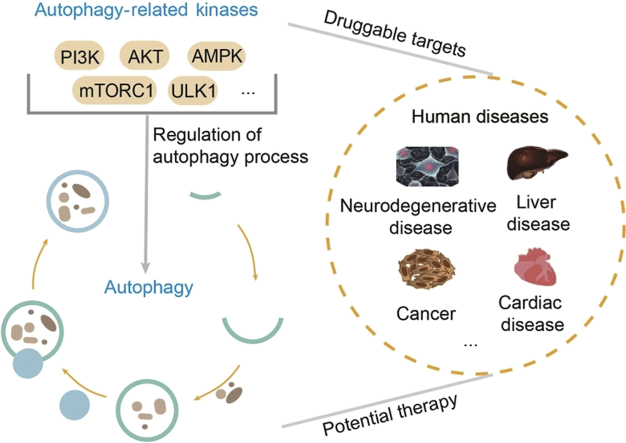
Graphical abstract
This review elaborates in detail the role of some autophagy-related kinase targets and phosphorylation in autophagy regulation, and further summarizes the application of small-molecule kinase inhibitors/activators for autophagy inhibition and induction. Understanding how these autophagy-related kinases regulate autophagy machinery is critical for the potential therapeutic application of autophagy.
Related collections
Most cited references106
- Record: found
- Abstract: found
- Article: not found
The protein kinase complement of the human genome.
- Record: found
- Abstract: found
- Article: not found
Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase.
- Record: found
- Abstract: found
- Article: not found