- Record: found
- Abstract: found
- Article: found
Different Chronic Stress Paradigms Converge on Endogenous TDP43 Cleavage and Aggregation
Read this article at
Abstract
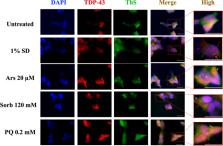
The TAR-DNA binding protein (TDP43) is a nuclear protein whose cytoplasmic inclusions are hallmarks of Amyotrophic Lateral Sclerosis (ALS). Acute stress in cells causes TDP43 mobilization to the cytoplasm and its aggregation through different routes. Although acute stress elicits a strong phenotype, is far from recapitulating the years-long aggregation process. We applied different chronic stress protocols and described TDP43 aggregation in a human neuroblastoma cell line by combining solubility assays, thioflavin-based microscopy and flow cytometry. This approach allowed us to detect, for the first time to our knowledge in vitro, the formation of 25 kDa C-terminal fragment of TDP43, a pathogenic hallmark of ALS. Our results indicate that chronic stress, compared to the more common acute stress paradigm, better recapitulates the cell biology of TDP43 proteinopathies. Moreover, we optimized a protocol for the detection of bona fide prions in living cells, suggesting that TDP43 may form amyloids as a stress response.
Related collections
Most cited references91
- Record: found
- Abstract: found
- Article: not found
Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis.
- Record: found
- Abstract: found
- Article: not found
Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases
- Record: found
- Abstract: found
- Article: not found