- Record: found
- Abstract: found
- Article: not found
Population genomics of post-vaccine changes in pneumococcal epidemiology
Read this article at
Abstract
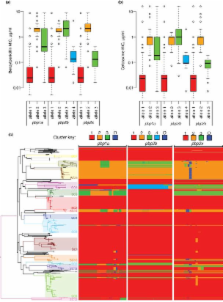
Whole genome sequencing of 616 asymptomatically carried pneumococci was used to study the impact of the 7-valent pneumococcal conjugate vaccine. Comparison of closely related isolates revealed the role of transformation in facilitating capsule switching to non-vaccine serotypes and the emergence of drug resistance. However, such recombination was found to occur at significantly different rates across the species, and the evolution of the population was primarily driven by changes in the frequency of distinct genotypes extant pre-vaccine. These alterations resulted in little overall effect on accessory genome composition at the population level, contrasting with the fall in pneumococcal disease rates after the vaccine’s introduction.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
The Bioperl toolkit: Perl modules for the life sciences.
- Record: found
- Abstract: not found
- Article: not found
R: A Language and Environment for Statistical Computing

- Record: found
- Abstract: found
- Article: found
