- Record: found
- Abstract: found
- Article: found
Production of ent-kaurene from lignocellulosic hydrolysate in Rhodosporidium toruloides
Read this article at
Abstract
Background
Rhodosporidium toruloides has emerged as a promising host for the production of bioproducts from lignocellulose, in part due to its ability to grow on lignocellulosic feedstocks, tolerate growth inhibitors, and co-utilize sugars and lignin-derived monomers. Ent-kaurene derivatives have a diverse range of potential applications from therapeutics to novel resin-based materials.
Results
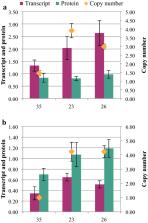
The Design, Build, Test, and Learn (DBTL) approach was employed to engineer production of the non-native diterpene ent-kaurene in R. toruloides. Following expression of kaurene synthase (KS) in R. toruloides in the first DBTL cycle, a key limitation appeared to be the availability of the diterpene precursor, geranylgeranyl diphosphate (GGPP). Further DBTL cycles were carried out to select an optimal GGPP synthase and to balance its expression with KS, requiring two of the strongest promoters in R. toruloides, ANT (adenine nucleotide translocase) and TEF1 (translational elongation factor 1) to drive expression of the KS from Gibberella fujikuroi and a mutant version of an FPP synthase from Gallus gallus that produces GGPP. Scale-up of cultivation in a 2 L bioreactor using a corn stover hydrolysate resulted in an ent-kaurene titer of 1.4 g/L.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
Skyline: an open source document editor for creating and analyzing targeted proteomics experiments.
- Record: found
- Abstract: not found
- Article: not found
High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture

- Record: found
- Abstract: found
- Article: found