- Record: found
- Abstract: found
- Article: found
Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways

Read this article at
SUMMARY
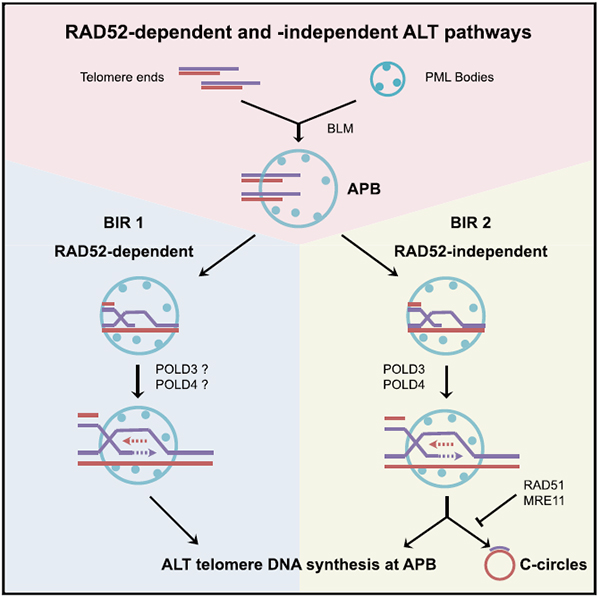
Alternative lengthening of telomeres (ALT) is a telomerase-independent but recombination-dependent pathway that maintains telomeres. Here, we describe an assay to visualize ALT-mediated telomeric DNA synthesis in ALT-associated PML bodies (APBs) without DNA-damaging agents or replication inhibitors. Using this assay, we find that ALT occurs through two distinct mechanisms. One of the ALT mechanisms requires RAD52, a protein implicated in break-induced DNA replication (BIR). We demonstrate that RAD52 directly promotes telomeric D-loop formation in vitro and is required for maintaining telomeres in ALT-positive cells. Unexpectedly, however, RAD52 is dispensable for C-circle formation, a hallmark of ALT. In RAD52-knockout ALT cells, C-circle formation and RAD52-independent ALT DNA synthesis gradually increase as telomeres are shortened, and these activities are dependent on BLM and BIR proteins POLD3 and POLD4. These results suggest that ALT occurs through a RAD52-dependent and a RAD52-independent BIR pathway, revealing the bifurcated framework and dynamic nature of this process.
Graphical Abstract
In Brief
Alternative lengthening of telomeres (ALT) is a telomerase-independent but recombination-dependent process that extends telomeres. Zhang et al. show that ALT is in fact a bifurcated pathway involving both RAD52-dependent and RAD52-independent break-induced DNA replication (BIR) in ALT-associated PML bodies (APBs), revealing an unexpected framework of the ALT pathway.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
Alternative lengthening of telomeres: models, mechanisms and implications.
- Record: found
- Abstract: found
- Article: not found
Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism.
- Record: found
- Abstract: found
- Article: not found