- Record: found
- Abstract: found
- Article: found
Bioinformatics and network pharmacology-based study to elucidate the multi-target pharmacological mechanism of the indigenous plants of Medina valley in treating HCV-related hepatocellular carcinoma
Read this article at
Abstract
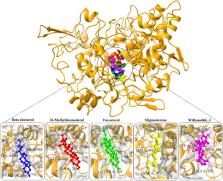
The incidence of Hepatocellular Carcinoma (HCC) in Saudi Arabia is not surprising given the relatively high prevalence of hepatitis C virus (HCV) infection. Hepatitis C is also common in Saudi Arabia with a prevalence rate of 1% to 3% of the population, which further increases the risk of HCC. The incidence of HCC has been increasing in recent years, with HCV-related HCC accounting for a significant proportion of cases. Traditional medicine has long been a part of Saudi Arabian culture, and many medicinal plants have been used for centuries to treat various ailments, including cancer. Following that, this study combines network pharmacology with bioinformatics approaches to potentially revolutionize HCV-related HCC treatment by identifying effective phytochemicals of indigenous plants of Medina valley. Eight indigenous plants including Rumex vesicarius, Withania somnifera, Rhazya stricta, Heliotropium arbainense, Asphodelus fistulosus, Pulicaria incise, Commicarpus grandiflorus, and Senna alexandrina, were selected for the initial screening of potential drug-like compounds. At first, the information related to active compounds of eight indigenous plants was retrieved from public databases and through literature review which was later combined with differentially expressed genes (DEGs) obtained through microarray datasets. Later, a compound-target genes-disease network was constructed which uncovered that kaempferol, rhazimol, beta-sitosterol, 12-Hydroxy-3-keto-bisnor-4-cholenic acid, 5-O-caffeoylquinic acid, 24-Methyldesmosterol, stigmasterone, fucosterol, and withanolide_J decisively contributed to the cell growth and proliferation by affecting ALB and PTGS2 proteins. Moreover, the molecular docking and Molecular Dynamic (MD) simulation of 20 ns well complemented the binding affinity of the compound and revealed strong stability of predicted compounds at the docked site. But the findings were not validated in actual patients, so further investigation is needed to confirm the potential use of selected medicinal plants towards HCV-related HC.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
Cytoscape: a software environment for integrated models of biomolecular interaction networks.
- Record: found
- Abstract: found
- Article: not found
