- Record: found
- Abstract: found
- Article: found
Genome-wide identification of a novel Na + transporter from Bienertia sinuspersici and overexpression of BsHKT1;2 improved salt tolerance in Brassica rapa
Read this article at
Abstract
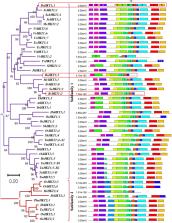
Salt stress is an ever-increasing stressor that affects both plants and humans. Therefore, developing strategies to limit the undesirable effects of salt stress is essential. Sodium ion exclusion is well known for its efficient salt-tolerance mechanism. The High-affinity K + Transporter (HKT) excludes excess Na + from the transpiration stream. This study identified and characterized the HKT protein family in Bienertia sinuspersici, a single-cell C 4 plant. The HKT and Salt Overly Sensitive 1 (SOS1) expression levels were examined in B. sinuspersici and Arabidopsis thaliana leaves under four different salt stress conditions: 0, 100, 200, and 300 mM NaCl. Furthermore, BsHKT1;2 was cloned, thereby producing stable transgenic Brassica rapa. Our results showed that, compared to A. thaliana as a glycophyte, the HKT family is expanded in B. sinuspersici as a halophyte with three paralogs. The phylogenetic analysis revealed three paralogs belonging to the HKT subfamily I. Out of three copies, the expression of BsHKT1;2 was higher in Bienertia under control and salt stress conditions than in A. thaliana. Stable transgenic plants overexpressing 35S::BsHKT1;2 showed higher salt tolerance than non-transgenic plants. Higher biomass and longer roots were observed in the transgenic plants under salt stress than in non-transgenic plants. This study demonstrates the evolutionary and functional differences in HKT proteins between glycophytes and halophytes and associates the role of BsHKT1;2 in imparting salt tolerance and productivity.
Related collections
Most cited references53

- Record: found
- Abstract: found
- Article: found
MEGA11: Molecular Evolutionary Genetics Analysis Version 11

- Record: found
- Abstract: found
- Article: found
MEME Suite: tools for motif discovery and searching
- Record: found
- Abstract: found
- Article: not found