- Record: found
- Abstract: found
- Article: not found
A SARS-CoV-2 Cluster in an Acute Care Hospital
Read this article at
Abstract
This study describes the detection, mitigation, and analysis of a large cluster of SARS-CoV-2 infections in an acute care hospital with mature infection control policies and discusses insights that may inform additional measures to protect patients and staff.
Abstract
This study describes the detection, mitigation, and analysis of a large cluster of SARS-CoV-2 infections in an acute care hospital with mature infection control policies and discusses insights that may inform additional measures to protect patients and staff.
Abstract
Background:
Little is known about clusters of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in acute care hospitals.
Objective:
To describe the detection, mitigation, and analysis of a large cluster of SARS-CoV-2 infections in an acute care hospital with mature infection control policies.
Intervention:
Close contacts of infected patients and staff were identified and tested every 3 days, patients on affected units were preemptively isolated and repeatedly tested, affected units were cleaned, room ventilation was measured, and specimens were sent for whole-genome sequencing. A case–control study was done to compare clinical interactions, personal protective equipment use, and breakroom and workroom practices in SARS-CoV-2–positive versus negative staff.
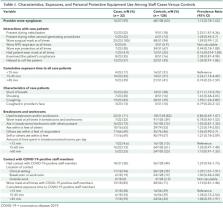
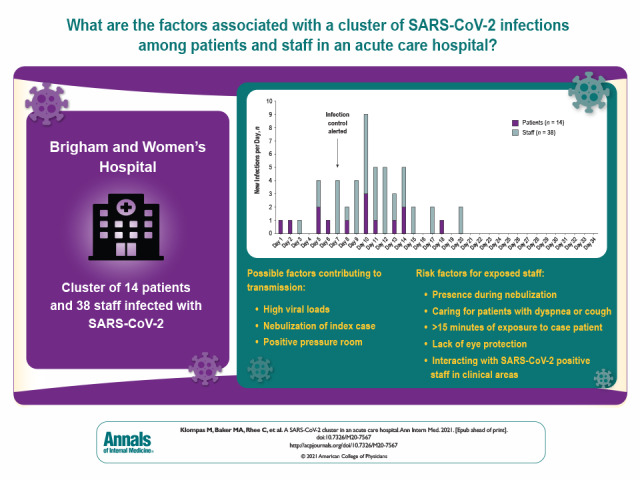
Results:
Fourteen patients and 38 staff members were included in the cluster per whole-genome sequencing and epidemiologic associations. The index case was a symptomatic patient in whom isolation was discontinued after 2 negative results on nasopharyngeal polymerase chain reaction testing. The patient subsequently infected multiple roommates and staff, who then infected others. Seven of 52 (13%) secondary infections were detected only on second or subsequent tests. Eight of 9 (89%) patients who shared rooms with potentially contagious patients became infected. Potential contributing factors included high viral loads, nebulization, and positive pressure in the index patient's room. Risk factors for transmission to staff included presence during nebulization, caring for patients with dyspnea or cough, lack of eye protection, at least 15 minutes of exposure to case patients, and interactions with SARS-CoV-2–positive staff in clinical areas. Whole-genome sequencing confirmed that 2 staff members were infected despite wearing surgical masks and eye protection.
Related collections
Most cited references35

- Record: found
- Abstract: found
- Article: found
Fast and accurate short read alignment with Burrows–Wheeler transform
- Record: found
- Abstract: found
- Article: not found
Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases
- Record: found
- Abstract: found
- Article: not found
