- Record: found
- Abstract: found
- Article: found
Identification of Immune Subtypes and Candidate mRNA Vaccine Antigens in Small Cell Lung Cancer
Read this article at
Abstract
Background
Immune checkpoint inhibitors (ICIs) have demonstrated promising outcomes in small cell lung cancer (SCLC), but not all patients benefit from it. Thus, developing precise treatments for SCLC is a particularly urgent need. In our study, we constructed a novel phenotype for SCLC based on immune signatures.
Methods
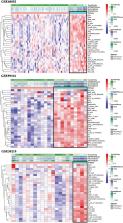
We clustered patients with SCLC hierarchically in 3 publicly available datasets according to the immune signatures. ESTIMATE and CIBERSORT algorithm were used to evaluate the components of the tumor microenvironment. Moreover, we identified potential mRNA vaccine antigens for patients with SCLC, and qRT-PCR were performed to detect the gene expression.
Results
We identified 2 SCLC subtypes and named Immunity High (Immunity_H) and Immunity Low (Immunity_L). Meanwhile, we obtained generally consistent results by analyzing different datasets, suggesting that this classification was reliable. Immunity_H contained the higher number of immune cells and a better prognosis compared to Immunity_L. Gene-set enrichment analysis revealed that several immune-related pathways such as cytokine-cytokine receptor interaction, programmed cell death-Ligand 1 expression and programmed cell death-1 checkpoint pathway in cancer were hyperactivated in the Immunity_H. However, most of the pathways enriched in the Immunity_L were not associated with immunity. Furthermore, we identified 5 potential mRNA vaccine antigens of SCLC (NEK2, NOL4, RALYL, SH3GL2, and ZIC2), and they were expressed higher in Immunity_L, it indicated that Immunity_L maybe more suitable for tumor vaccine development.
Abstract
Immune checkpoint inhibitors (ICIs) have shown promising outcomes in small cell lung cancer (SCLC), but not all patients benefit from ICIs therapy. Developing precise treatments for SCLC is a particularly urgent need. This article describes a novel phenotype for SCLC based on immune signatures.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries

- Record: found
- Abstract: found
- Article: found
GSVA: gene set variation analysis for microarray and RNA-Seq data
- Record: found
- Abstract: found
- Article: not found