- Record: found
- Abstract: found
- Article: found
Prospective clinical sequencing of 1016 Chinese prostate cancer patients: uncovering genomic characterization and race disparity
Read this article at
Abstract
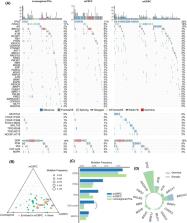
Although there is a well‐known disparity in prostate cancer (PC) incidence and mortality between Chinese and Western patients, the underlying genomic differences have been investigated only sparsely. This clinicogenomic study was conducted to reveal the genomic mutations contributing to the PC disparity across ethnicities and investigate the mutational profile of Chinese PC patients. A total of 1016 Chinese PC patients were prospectively enrolled and subjected to targeted sequencing, resulting in usable sequencing data for 41 genes from 859 patients. Genomic data retrieved from The Cancer Genome Atlas (TCGA; locoregional PC), Memorial Sloan Kettering Cancer Center [MSKCC; metastatic castration‐sensitive PC (mCSPC)], and Stand Up To Cancer [SU2C; metastatic castration‐resistant PC (mCRPC)] cohorts were used as comparators representing Western men. Genomic mutations were analyzed using an integrated bioinformatic strategy. A comparison of the disease stages revealed that mutations in tumor protein 53 ( TP53), androgen receptor ( AR), forkhead box A1 ( FOXA1), and genes involved in the cell cycle pathway were enriched in mCRPC. Mutations in adenomatous polyposis coli ( APC) gene were found to be more prevalent in patients with visceral metastasis. Genomic differences between Western and Chinese men were mainly observed in castration‐sensitive PC, with tumors from Chinese men having more FOXA1 (11.4% vs. 4.2%) but fewer TP53 (4.8% vs. 13%) mutations in locoregional PC and harboring fewer TP53 (11% vs. 29.2%), phosphatase and tensin homolog ( PTEN; 2.5% vs. 10.3%), and APC (1.7% vs. 7.4%) mutations in the mCSPC stage than those of Western men. Patients of both ethnicities with mCRPC had similar mutational spectra. Furthermore, FOXA1 class‐2 was less common than FOXA1 class‐1 and showed no enrichment in metastasis, contrary to the findings in the Western cohort. Our study provides a valuable resource for a better understanding of PC in China and reveals the genomic alterations associated with PC disparity across races.
Abstract
The authors sequenced the genomic DNA from 1016 Chinese prostate cancer patients. Mutations in TP53, AR, and FOXA1 were enriched in metastatic castration‐resistant prostate cancers (mCRPCs), and APC mutations were enriched in liver metastasis. The authors also found notable genomic difference between tumors in Chinese and Western men, with more FOXA1 but fewer TP53, PTEN, and APC mutations in Chinese men.
Related collections
Most cited references65
- Record: found
- Abstract: found
- Article: not found
Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries

- Record: found
- Abstract: found
- Article: found
