- Record: found
- Abstract: found
- Article: not found
Immunogenic neoantigens derived from gene fusions stimulate T cell responses
Read this article at
Abstract
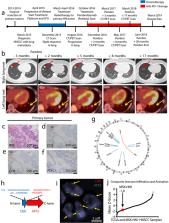
Anti-tumor immunity is driven by self vs. non-self discrimination. Many immunotherapeutic approaches to cancer have taken advantage of tumor neoantigens derived from somatic mutations. Here, we demonstrate that gene fusions are a source of immunogenic neoantigens that can mediate responses to immunotherapy. We identified an exceptional responder with metastatic head and neck cancer who experienced a complete response to immune checkpoint inhibitor therapy, despite a low mutational load and minimal pre-treatment immune infiltration in the tumor. Using whole genome sequencing (WGS) and RNA sequencing (RNA-seq), we identified a novel gene fusion, and demonstrated that it produces a neoantigen that can specifically elicit a host cytotoxic T cell response. In a cohort of head and neck tumors with low mutation burden, minimal immune infiltration, and prevalent gene fusions, we also identified gene fusion-derived neoantigens that generate cytotoxic T cell responses. Finally, analyzing additional datasets of fusion-positive cancers, including checkpoint inhibitor-treated tumors, we found evidence of immune surveillance resulting in negative selective pressure against gene fusion-derived neoantigens. These findings highlight an important class of tumor-specific antigens, and have implications for targeting gene fusion events in cancers that would otherwise be less poised for response to immunotherapy, including cancers with low mutational load and minimal immune infiltration.
Related collections
Most cited references16
- Record: found
- Abstract: found
- Article: not found
Identifying specificity groups in the T cell receptor repertoire
- Record: found
- Abstract: found
- Article: not found
Immunogenicity of somatic mutations in human gastrointestinal cancers.
- Record: found
- Abstract: found
- Article: not found
