- Record: found
- Abstract: found
- Article: found
Centriole and Golgi microtubule nucleation are dispensable for the migration of human neutrophil-like cells
Read this article at
Abstract
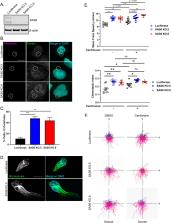
Neutrophils migrate in response to chemoattractants to mediate host defense. Chemoattractants drive rapid intracellular cytoskeletal rearrangements including the radiation of microtubules from the microtubule-organizing center (MTOC) toward the rear of polarized neutrophils. Microtubules regulate neutrophil polarity and motility, but little is known about the specific role of MTOCs. To characterize the role of MTOCs on neutrophil motility, we depleted centrioles in a well-established neutrophil-like cell line. Surprisingly, both chemical and genetic centriole depletion increased neutrophil speed and chemotactic motility, suggesting an inhibitory role for centrioles during directed migration. We also found that depletion of both centrioles and GM130-mediated Golgi microtubule nucleation did not impair neutrophil directed migration. Taken together, our findings demonstrate an inhibitory role for centrioles and a resilient MTOC system in motile human neutrophil-like cells.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: found
Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR
- Record: found
- Abstract: found
- Article: not found
High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities.
- Record: found
- Abstract: found
- Article: not found