- Record: found
- Abstract: found
- Article: found
Unraveling the intercellular communication disruption and key pathways in Alzheimer’s disease: An integrative study of single-nucleus transcriptomes and genetic association
Read this article at
Abstract
Background
Recently, single-nucleus RNA-seq (snRNA-seq) analyses have revealed important cellular and functional features of Alzheimer’s disease (AD), a prevalent neurodegenerative disease. However, our knowledge regarding intercellular communication mediated by dysregulated ligand-receptor (LR) interactions remains very limited in AD brains.
Methods
We systematically assessed the intercellular communication networks by using a discovery snRNA-seq dataset comprising 69,499 nuclei from 48 human postmortem prefrontal cortex (PFC) samples. We replicated the findings using an independent snRNA-seq dataset of 56,440 nuclei from 18 PFC samples. By integrating genetic signals from AD genome-wide association studies (GWAS) summary statistics and whole genome sequencing (WGS) data, we prioritized AD-associated Gene Ontology (GO) terms containing dysregulated LR interactions. We further explored drug repurposing for the prioritized LR pairs using the Therapeutic Targets Database.
Results
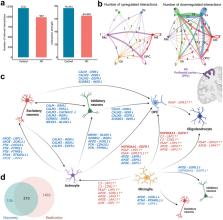
We identified 316 dysregulated LR interactions across six major cell types in AD PFC, of which 210 pairs were replicated. Among the replicated LR signals, we found globally downregulated communications in astrocytes-to-neurons signaling axis, characterized, for instance, by the downregulation of APOE-related and Calmodulin (CALM)-related LR interactions and their potential regulatory connections to target genes. Pathway analyses revealed 60 GO terms significantly linked to AD, highlighting Biological Processes such as ‘amyloid precursor protein processing’ and ‘ion transmembrane transport’, among others. We prioritized several drug repurposing candidates, such as cromoglicate, targeting the identified dysregulated LR pairs.
Related collections
Most cited references85
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles

- Record: found
- Abstract: found
- Article: found