- Record: found
- Abstract: found
- Article: found
Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients
Read this article at
Abstract
Background
The SARS-CoV-2 pandemic is currently leading to increasing numbers of COVID-19 patients all over the world. Clinical presentations range from asymptomatic, mild respiratory tract infection, to severe cases with acute respiratory distress syndrome, respiratory failure, and death. Reports on a dysregulated immune system in the severe cases call for a better characterization and understanding of the changes in the immune system.
Methods
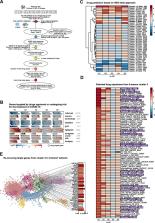
In order to dissect COVID-19-driven immune host responses, we performed RNA-seq of whole blood cell transcriptomes and granulocyte preparations from mild and severe COVID-19 patients and analyzed the data using a combination of conventional and data-driven co-expression analysis. Additionally, publicly available data was used to show the distinction from COVID-19 to other diseases. Reverse drug target prediction was used to identify known or novel drug candidates based on finding from data-driven findings.
Results
Here, we profiled whole blood transcriptomes of 39 COVID-19 patients and 10 control donors enabling a data-driven stratification based on molecular phenotype. Neutrophil activation-associated signatures were prominently enriched in severe patient groups, which was corroborated in whole blood transcriptomes from an independent second cohort of 30 as well as in granulocyte samples from a third cohort of 16 COVID-19 patients (44 samples). Comparison of COVID-19 blood transcriptomes with those of a collection of over 3100 samples derived from 12 different viral infections, inflammatory diseases, and independent control samples revealed highly specific transcriptome signatures for COVID-19. Further, stratified transcriptomes predicted patient subgroup-specific drug candidates targeting the dysregulated systemic immune response of the host.
Conclusions
Our study provides novel insights in the distinct molecular subgroups or phenotypes that are not simply explained by clinical parameters. We show that whole blood transcriptomes are extremely informative for COVID-19 since they capture granulocytes which are major drivers of disease severity.
Related collections
Most cited references125

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China
- Record: found
- Abstract: found
- Article: not found