- Record: found
- Abstract: found
- Article: found
The OceanDNA MAG catalog contains over 50,000 prokaryotic genomes originated from various marine environments
Read this article at
Abstract
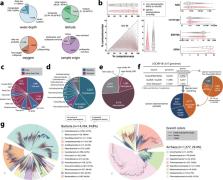
Marine microorganisms are immensely diverse and play fundamental roles in global geochemical cycling. Recent metagenome-assembled genome studies, with particular attention to large-scale projects such as Tara Oceans, have expanded the genomic repertoire of marine microorganisms. However, published marine metagenome data is still underexplored. We collected 2,057 marine metagenomes covering various marine environments and developed a new genome reconstruction pipeline. We reconstructed 52,325 qualified genomes composed of 8,466 prokaryotic species-level clusters spanning 59 phyla, including genomes from the deep-sea characterized as deeper than 1,000 m (n = 3,337), low-oxygen zones of <90 μmol O2 per kg water (n = 7,884), and polar regions (n = 7,752). Novelty evaluation using a genome taxonomy database shows that 6,256 species (73.9%) are novel and include genomes of high taxonomic novelty, such as new class candidates. These genomes collectively expanded the known phylogenetic diversity of marine prokaryotes by 34.2%, and the species representatives cover 26.5–42.0% of prokaryote-enriched metagenomes. Thoroughly leveraging accumulated metagenomic data, this genome resource, named the OceanDNA MAG catalog, illuminates uncharacterized marine microbial ‘dark matter’ lineages.
Abstract
Related collections
Most cited references86

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.
- Record: found
- Abstract: found
- Article: not found