- Record: found
- Abstract: found
- Article: found
Differentiation between thyroid‐associated orbitopathy and Graves’ disease by iTRAQ‐based quantitative proteomic analysis

Read this article at
Abstract
Graves’ ophthalmopathy, also known as thyroid‐associated orbitopathy (TAO), is the most common inflammatory eye disease in adults. The most common etiology for TAO is Graves’ disease (GD); however, proteomic research focusing on differences between GD and TAO is limited. This study aimed to identify differentially expressed proteins between thyroid‐associated orbitopathy (TAO) and GD. Furthermore, we sought to explore the pathogenesis of TAO and elucidate the differentiation process via specific markers. Serum samples of three patients with TAO, GD, and healthy controls, respectively, were collected. These samples were measured using the iTRAQ technique coupled with mass spectrometry. Differentially expressed proteins in TAO and GD were identified by proteomics; 3172 quantified proteins were identified. Compared with TAO, we identified 110 differential proteins (27 proteins were upregulated and 83 were downregulated). In addition, these differentially expressed proteins were closely associated with cellular processes, metabolic processes, macromolecular complexes, signal transduction, and the immune system. The corresponding functions were protein, calcium ion, and nucleic acid binding. Among the differential proteins, MYH11, P4HB, and C4A were markedly upregulated in TAO patients and have been reported to participate in apoptosis, autophagy, the inflammatory response, and the immune system. A protein–protein interaction network analysis was performed. Proteomics demonstrated valuable large‐scale protein‐related information for expounding the pathogenic mechanism underlying TAO. This research provides new insights and potential targets for studying GD with TAO.
Abstract
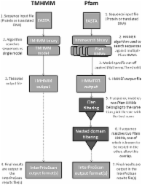
Serum samples were collected from patients with TAO and Graves’ disease (GD), and healthy controls (three cases in each group). After extraction and quantification, the obtained proteins were treated with ITRAQ reagent and then analyzed using LC‐MS/MS. Finally, the identified differentially expressed proteins in TAO and GD were subjected to bioinformatics analysis, including GO analysis, KEGG analysis, and PPI analysis.
Related collections
Most cited references39

- Record: found
- Abstract: found
- Article: found
Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists
- Record: found
- Abstract: found
- Article: found
InterProScan 5: genome-scale protein function classification
- Record: found
- Abstract: found
- Article: not found
