- Record: found
- Abstract: found
- Article: found
Enhanced Virus Detection and Metagenomic Sequencing in Patients with Meningitis and Encephalitis
Read this article at
ABSTRACT
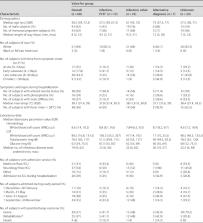
Meningitis and encephalitis are leading causes of central nervous system (CNS) disease and often result in severe neurological compromise or death. Traditional diagnostic workflows largely rely on pathogen-specific tests, sometimes over days to weeks, whereas metagenomic next-generation sequencing (mNGS) profiles all nucleic acid in a sample. In this single-center, prospective study, 68 hospitalized patients with known ( n = 44) or suspected ( n = 24) CNS infections underwent mNGS from RNA and DNA to identify potential pathogens and also targeted sequencing of viruses using hybrid capture. Using a computational metagenomic classification pipeline based on KrakenUniq and BLAST, we detected pathogen nucleic acid in cerebrospinal fluid (CSF) from 22 subjects, 3 of whom had no clinical diagnosis by routine workup. Among subjects diagnosed with infection by serology and/or peripheral samples, we demonstrated the utility of mNGS to detect pathogen nucleic acid in CSF, importantly for the Ixodes scapularis tick-borne pathogens Powassan virus, Borrelia burgdorferi, and Anaplasma phagocytophilum. We also evaluated two methods to enhance the detection of viral nucleic acid, hybrid capture and methylated DNA depletion. Hybrid capture nearly universally increased viral read recovery. Although results for methylated DNA depletion were mixed, it allowed the detection of varicella-zoster virus DNA in two samples that were negative by standard mNGS. Overall, mNGS is a promising approach that can test for multiple pathogens simultaneously, with efficacy similar to that of pathogen-specific tests, and can uncover geographically relevant infectious CNS disease, such as tick-borne infections in New England. With further laboratory and computational enhancements, mNGS may become a mainstay of workup for encephalitis and meningitis.
Related collections
Most cited references57

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found
Basic local alignment search tool.

- Record: found
- Abstract: found
- Article: found