- Record: found
- Abstract: found
- Article: not found
Classification of COVID-19 from chest x-ray images using deep features and correlation coefficient
Read this article at
Abstract
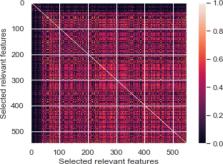
COVID-19 is a viral disease that in the form of a pandemic has spread in the entire world, causing a severe impact on people’s well being. In fighting against this deadly disease, a pivotal step can prove to be an effective screening and diagnosing step to treat infected patients. This can be made possible through the use of chest X-ray images. Early detection using the chest X-ray images can prove to be a key solution in fighting COVID-19. Many computer-aided diagnostic (CAD) techniques have sprung up to aid radiologists and provide them a secondary suggestion for the same. In this study, we have proposed the notion of Pearson Correlation Coefficient (PCC) along with variance thresholding to optimally reduce the feature space of extracted features from the conventional deep learning architectures, ResNet152 and GoogLeNet. Further, these features are classified using machine learning (ML) predictive classifiers for multi-class classification among COVID-19, Pneumonia and Normal. The proposed model is validated and tested on publicly available COVID-19 and Pneumonia and Normal dataset containing an extensive set of 768 images of COVID-19 with 5216 training images of Pneumonia and Normal patients. Experimental results reveal that the proposed model outperforms other previous related works. While the achieved results are encouraging, further analysis on the COVID-19 images can prove to be more reliable for effective classification.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study
- Record: found
- Abstract: found
- Article: not found
