- Record: found
- Abstract: found
- Article: found
The Mitochondrial Protein VDAC1 at the Crossroads of Cancer Cell Metabolism: The Epigenetic Link
Read this article at
Abstract
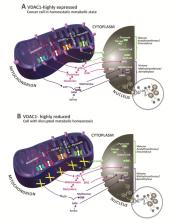
Carcinogenesis is a complicated process that involves the deregulation of epigenetics, resulting in cellular transformational events, such as proliferation, differentiation, and metastasis. Most chromatin-modifying enzymes utilize metabolites as co-factors or substrates and thus are directly dependent on such metabolites as acetyl-coenzyme A, S-adenosylmethionine, and NAD+. Here, we show that using specific siRNA to deplete a tumor of VDAC1 not only led to reprograming of the cancer cell metabolism but also altered several epigenetic-related enzymes and factors. VDAC1, in the outer mitochondrial membrane, controls metabolic cross-talk between the mitochondria and the rest of the cell, thus regulating the metabolic and energetic functions of mitochondria, and has been implicated in apoptotic-relevant events. We previously demonstrated that silencing VDAC1 expression in glioblastoma (GBM) U-87MG cell-derived tumors, resulted in reprogramed metabolism leading to inhibited tumor growth, angiogenesis, epithelial–mesenchymal transition and invasiveness, and elimination of cancer stem cells, while promoting the differentiation of residual tumor cells into neuronal-like cells. These VDAC1 depletion-mediated effects involved alterations in transcription factors regulating signaling pathways associated with cancer hallmarks. As the epigenome is sensitive to cellular metabolism, this study was designed to assess whether depleting VDAC1 affects the metabolism–epigenetics axis. Using DNA microarrays, q-PCR, and specific antibodies, we analyzed the effects of si-VDAC1 treatment of U-87MG-derived tumors on histone modifications and epigenetic-related enzyme expression levels, as well as the methylation and acetylation state, to uncover any alterations in epigenetic properties. Our results demonstrate that metabolic rewiring of GBM via VDAC1 depletion affects epigenetic modifications, and strongly support the presence of an interplay between metabolism and epigenetics.
Related collections
Most cited references69
- Record: found
- Abstract: found
- Article: not found
A unique chromatin signature uncovers early developmental enhancers in humans.
- Record: found
- Abstract: found
- Article: not found
Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1.
- Record: found
- Abstract: found
- Article: not found