- Record: found
- Abstract: found
- Article: found
ABI5–FLZ13 module transcriptionally represses growth-related genes to delay seed germination in response to ABA
Read this article at
Abstract
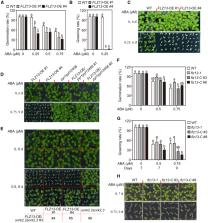
The bZIP transcription factor ABSCISIC ACID INSENSITIVE5 (ABI5) is a master regulator of seed germination and post-germinative growth in response to abscisic acid (ABA), but the detailed molecular mechanism by which it represses plant growth remains unclear. In this study, we used proximity labeling to map the neighboring proteome of ABI5 and identified FCS-LIKE ZINC FINGER PROTEIN 13 (FLZ13) as a novel ABI5 interaction partner. Phenotypic analysis of flz13 mutants and FLZ13-overexpressing lines demonstrated that FLZ13 acts as a positive regulator of ABA signaling. Transcriptomic analysis revealed that both FLZ13 and ABI5 downregulate the expression of ABA-repressed and growth-related genes involved in chlorophyll biosynthesis, photosynthesis, and cell wall organization, thereby repressing seed germination and seedling establishment in response to ABA. Further genetic analysis showed that FLZ13 and ABI5 function together to regulate seed germination. Collectively, our findings reveal a previously uncharacterized transcriptional regulatory mechanism by which ABA mediates inhibition of seed germination and seedling establishment.
Abstract
FLZ13 physically interacts with ABI5 to downregulate the expression of ABA-repressed and growth-related genes, thereby inhibiting seed germination and seedling establishment in response to ABA.
Related collections
Most cited references78
- Record: found
- Abstract: found
- Article: not found
Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources.

- Record: found
- Abstract: found
- Article: found
Metascape provides a biologist-oriented resource for the analysis of systems-level datasets
- Record: found
- Abstract: found
- Article: not found