- Record: found
- Abstract: found
- Article: found
The sea urchin ( Strongylocentrotus purpuratus) test and spine proteomes
Read this article at
Abstract
Background
The organic matrix of biominerals plays an important role in biomineral formation and in determining biomineral properties. However, most components of biomineral matrices remain unknown at present. In sea urchin, which is an important model organism for developmental biology and biomineralization, only few matrix components have been identified and characterized at the protein level. The recent publication of the Strongylocentrotus purpuratus genome sequence rendered possible not only the identification of possible matrix proteins at the gene level, but also the direct identification of proteins contained in matrices of skeletal elements by in-depth, high-accuracy, proteomic analysis.
Results
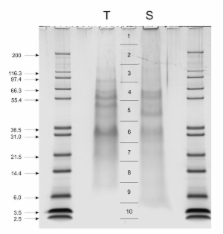
We identified 110 proteins as components of sea urchin test and spine organic matrix. Fourty of these proteins occurred in both compartments while others were unique to their respective compartment. More than 95% of the proteins were detected in sea urchin skeletal matrices for the first time. The most abundant protein in both matrices was the previously characterized spicule matrix protein SM50, but at least eight other members of this group, many of them only known as conceptual translation products previously, were identified by mass spectrometric sequence analysis of peptides derived from in vitro matrix degradation. The matrices also contained proteins implicated in biomineralization processes previously by inhibition studies using antibodies or specific enzyme inhibitors, such as matrix metalloproteases and members of the mesenchyme-specific MSP130 family. Other components were carbonic anhydrase, collagens, echinonectin, a α2-macroglobulin-like protein and several proteins containing scavenger receptor cysteine-rich domains. A few possible signal transduction pathway components, such as GTP-binding proteins, a semaphorin and a possible tyrosine kinase were also identified.
Conclusion
This report presents the most comprehensive list of sea urchin skeletal matrix proteins available at present. The complex mixture of proteins identified in matrices of the sea urchin skeleton may reflect many different aspects of the mineralization process. Because LC-MS/MS-based methods directly measures peptides our results validate many predicted genes and confirm the existence of the corresponding proteins. Considering the many newly identified matrix proteins, this proteomic study may serve as a road map for the further exploration of biomineralization processes in an important model organism.
Related collections
Most cited references53
- Record: found
- Abstract: not found
- Article: not found
Design strategies in mineralized biological materials

- Record: found
- Abstract: found
- Article: found
The 20 years of PROSITE
- Record: found
- Abstract: not found
- Article: not found