- Record: found
- Abstract: found
- Article: found
GENCODE reference annotation for the human and mouse genomes
Read this article at
Abstract
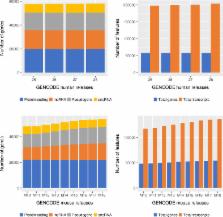
The accurate identification and description of the genes in the human and mouse genomes is a fundamental requirement for high quality analysis of data informing both genome biology and clinical genomics. Over the last 15 years, the GENCODE consortium has been producing reference quality gene annotations to provide this foundational resource. The GENCODE consortium includes both experimental and computational biology groups who work together to improve and extend the GENCODE gene annotation. Specifically, we generate primary data, create bioinformatics tools and provide analysis to support the work of expert manual gene annotators and automated gene annotation pipelines. In addition, manual and computational annotation workflows use any and all publicly available data and analysis, along with the research literature to identify and characterise gene loci to the highest standard. GENCODE gene annotations are accessible via the Ensembl and UCSC Genome Browsers, the Ensembl FTP site, Ensembl Biomart, Ensembl Perl and REST APIs as well as https://www.gencodegenes.org.
Related collections
Most cited references21

- Record: found
- Abstract: found
- Article: found
PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions
- Record: found
- Abstract: found
- Article: not found
Assessment of transcript reconstruction methods for RNA-seq
- Record: found
- Abstract: found
- Article: not found