- Record: found
- Abstract: found
- Article: found
The beagle dog MicroRNA tissue atlas: identifying translatable biomarkers of organ toxicity
Read this article at
Abstract
Background
MicroRNAs (miRNA) are varied in length, under 25 nucleotides, single-stranded noncoding RNA that regulate post-transcriptional gene expression via translational repression or mRNA degradation. Elevated levels of miRNAs can be detected in systemic circulation after tissue injury, suggesting that miRNAs are released following cellular damage. Because of their remarkable stability, ease of detection in biofluids, and tissue specific expression patterns, miRNAs have the potential to be specific biomarkers of organ injury. The identification of miRNA biomarkers requires a systematic approach: 1) determine the miRNA tissue expression profiles within a mammalian species via next generation sequencing; 2) identify enriched and/or specific miRNA expression within organs of toxicologic interest, and 3) in vivo validation with tissue-specific toxicants. While miRNA tissue expression has been reported in rodents and humans, little data exists on miRNA tissue expression in the dog, a relevant toxicology species. The generation and evaluation of the first dog miRNA tissue atlas is described here.
Results
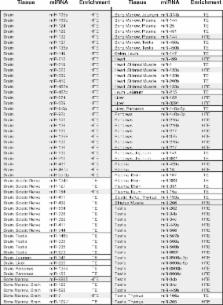
Analysis of 16 tissues from five male beagle dogs identified 106 tissue enriched miRNAs, 60 of which were highly enriched in a single organ, and thus may serve as biomarkers of organ injury. A proof of concept study in dogs dosed with hepatotoxicants evaluated a qPCR panel of 15 tissue enriched miRNAs specific to liver, heart, skeletal muscle, pancreas, testes, and brain. Dogs with elevated serum levels of miR-122 and miR-885 had a correlative increase of alanine aminotransferase, and microscopic analysis confirmed liver damage. Other non-liver enriched miRNAs included in the screening panel were unaffected. Eli Lilly authors created a complimentary Sprague Dawely rat miRNA tissue atlas and demonstrated increased pancreas enriched miRNA levels in circulation, following caerulein administration in rat and dog.
Conclusion
The dog miRNA tissue atlas provides a resource for biomarker discovery and can be further mined with refinement of dog genome annotation. The 60 highly enriched tissue miRNAs identified within the dog miRNA tissue atlas could serve as diagnostic biomarkers and will require further validation by in vivo correlation to histopathology. Once validated, these tissue enriched miRNAs could be combined into a powerful qPCR screening panel to identify organ toxicity during early drug development.
Related collections
Most cited references39

- Record: found
- Abstract: found
- Article: found
Characterization of microRNA expression profiles in normal human tissues
- Record: found
- Abstract: found
- Article: not found
Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases.

- Record: found
- Abstract: found
- Article: found