- Record: found
- Abstract: found
- Article: found
High-throughput analysis of the human thymic Vδ1 + T cell receptor repertoire
Read this article at
Abstract
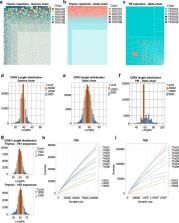
γδ T cells are a relatively rare subset of lymphocytes in the human peripheral blood, but they play important roles at the interface between the innate and the adaptive immune systems. The γδ T cell lineage is characterized by a signature γδ T cell receptor (γδTCR) that displays extensive sequence variability originated by DNA rearrangement of the corresponding V(D)J loci. Human γδ T cells comprise Vγ9Vδ2 T cells, the major subset in the peripheral blood; and Vδ1 + T cells, the predominant subpopulation in the post-natal thymus and in peripheral tissues. While less studied, Vδ1 + T cells recently gathered significant attention due to their anti-cancer and anti-viral activities. In this study we applied next-generation sequencing (NGS) to analyse the γδTCR repertoire of highly (FACS-)purified Vδ1 + T cells from human thymic biopsies. Our analysis reveals unsuspected aspects of thymically rearranged and expressed (at the mRNA level) TRG and TRD genes, thus constituting a data resource that qualifies previous conclusions on the TCR repertoire of γδ T cells developing in the human thymus.
Abstract
| Design Type(s) | transcription profiling design • cellular data analysis objective |
| Measurement Type(s) | gamma delta T-lymphocyte |
| Technology Type(s) | RNA sequencing |
| Factor Type(s) | sex • gene |
| Sample Characteristic(s) | Homo sapiens • thymus |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Related collections
Most cited references16
- Record: found
- Abstract: found
- Article: not found
γδ T cells in cancer.

- Record: found
- Abstract: found
- Article: found