- Record: found
- Abstract: found
- Article: found
Aberrant Nuclear Export of circNCOR1 Underlies SMAD7-Mediated Lymph Node Metastasis of Bladder Cancer

Read this article at
Abstract
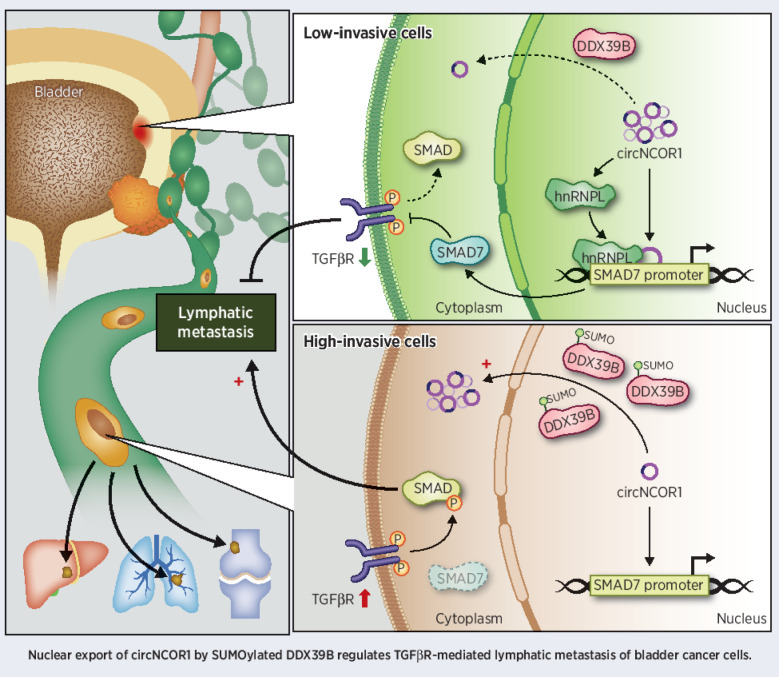
This study identifies the novel intron-retained circNCOR1 and elucidates a SUMOylation-mediated DDX39B–circNCOR1–SMAD7 axis that regulates lymph node metastasis of bladder cancer.
Abstract
Circular RNAs (circRNA) containing retained introns are normally sequestered in the nucleus. Dysregulation of cellular homeostasis can drive their nuclear export, which may be involved in cancer metastasis. However, the mechanism underlying circRNA nuclear export and its role in lymph node (LN) metastasis of bladder cancer remain unclear. Here, we identify an intron-retained circRNA, circNCOR1, that is significantly downregulated in LN metastatic bladder cancer and is negatively associated with poor prognosis of patients. Overexpression of circNCOR1 inhibited lymphangiogenesis and LN metastasis of bladder cancer in vitro and in vivo. Nuclear circNCOR1 epigenetically promoted SMAD7 transcription by increasing heterogeneous nuclear ribonucleoprotein L (hnRNPL)–induced H3K9 acetylation in the SMAD7 promoter, leading to inhibition of the TGFβ-SMAD signaling pathway. Nuclear retention of circNCOR1 was regulated by small ubiquitin-like modifier (SUMO)ylation of DDX39B, an essential regulatory factor responsible for circRNA nuclear-cytoplasmic transport. Reduced SUMO2 binding to DDX39B markedly increased circNCOR1 retention in the nucleus to suppress bladder cancer LN metastasis. By contrast, SUMOylated DDX39B activated nuclear export of circNCOR1, impairing the suppressive role of circNCOR1 on TGFβ-SMAD cascade activation and bladder cancer LN metastasis. In patient-derived xenograft (PDX) models, overexpression of circNCOR1 and inhibition of TGFβ signaling significantly repressed tumor growth and LN metastasis. This study highlights SUMOylation-induced nuclear export of circNCOR1 as a key event regulating TGFβ-SMAD signaling and bladder cancer lymphangiogenesis, thus supporting circNCOR1 as a novel therapeutic agent for patients with LN metastatic bladder cancer.
Graphical Abstract
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
Exon-intron circular RNAs regulate transcription in the nucleus.
- Record: found
- Abstract: not found
- Article: not found
The expanding regulatory mechanisms and cellular functions of circular RNAs

- Record: found
- Abstract: found
- Article: found