- Record: found
- Abstract: found
- Article: found
Bioinformatics analyses of potential ACLF biological mechanisms and identification of immune-related hub genes and vital miRNAs
Read this article at
Abstract
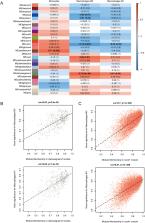
Acute-on-chronic liver failure (ACLF) is a critical and refractory disease and a hepatic disorder accompanied by immune dysfunction. Thus, it is essential to explore key immune-related genes of ACLF and investigate its mechanisms. We used two public datasets (GSE142255 and GSE168048) to perform various bioinformatics analyses, including WGCNA, CIBERSORT, and GSEA. We also constructed an ACLF immune-related protein–protein interaction (PPI) network to obtain hub differentially expressed genes (DEGs) and predict corresponding miRNAs. Finally, an ACLF rat model was established to verify the results. A total of 388 DEGs were identified in ACLF, including 162 upregulated and 226 downregulated genes. The enrichment analyses revealed that these DEGs were mainly involved in inflammatory-immune responses and biosynthetic metabolic pathways. Twenty-eight gene modules were obtained using WGCNA and the coral1 and darkseagreen4 modules were highly correlated with M1 macrophage polarization. As a result, 10 hub genes and 2 miRNAs were identified to be significantly altered in ACLF. The bioinformatics analyses of the two datasets presented valuable insights into the pathogenesis and screening of hub genes of ACLF. These results might contribute to a better understanding of the potential molecular mechanisms of ACLF. Finally, further studies are required to validate our current findings.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles

- Record: found
- Abstract: found
- Article: found
WGCNA: an R package for weighted correlation network analysis
- Record: found
- Abstract: found
- Article: not found