- Record: found
- Abstract: found
- Article: found
The Role of lncRNAs in Gene Expression Regulation through mRNA Stabilization
Read this article at
Abstract
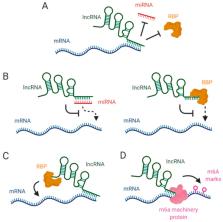
mRNA stability influences gene expression and translation in almost all living organisms, and the levels of mRNA molecules in the cell are determined by a balance between production and decay. Maintaining an accurate balance is crucial for the correct function of a wide variety of biological processes and to maintain an appropriate cellular homeostasis. Long non-coding RNAs (lncRNAs) have been shown to participate in the regulation of gene expression through different molecular mechanisms, including mRNA stabilization. In this review we provide an overview on the molecular mechanisms by which lncRNAs modulate mRNA stability and decay. We focus on how lncRNAs interact with RNA binding proteins and microRNAs to avoid mRNA degradation, and also on how lncRNAs modulate epitranscriptomic marks that directly impact on mRNA stability.
Related collections
Most cited references115
- Record: found
- Abstract: found
- Article: not found
A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language?
- Record: found
- Abstract: found
- Article: not found
m6A-dependent regulation of messenger RNA stability
- Record: found
- Abstract: found
- Article: not found