- Record: found
- Abstract: found
- Article: found
Investigation of phytochemicals isolated from selected Saudi medicinal plants as natural inhibitors of SARS CoV-2 main protease: In vitro , molecular docking and simulation analysis

Read this article at
Graphical abstract
Abstract
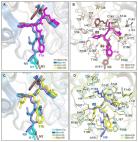
The escalation of many coronavirus variants accompanied by the lack of an effective cure has motivated the hunt for effective antiviral medicines. In this regard, 18 Saudi Arabian medicinal plants were evaluated for SARS CoV-2 main protease (Mpro) inhibition activity. Among them, Terminalia brownii and Acacia asak alcoholic extracts exhibited significant Mpro inhibition, with inhibition rates of 95.3 % and 95.2 %, respectively, at a concentration of 100 µg/mL. Bioassay-guided phytochemical study for the most active n-butanol fraction of T. brownii led to identification of eleven compounds, including two phenolic acids ( 1, and 2), seven hydrolysable tannins ( 3– 10), and one flavonoid ( 11) as well as four flavonoids from A. asak ( 12– 15). The structures of the isolated compounds were established using various spectroscopic techniques and comparison with known compounds. To investigate the chemical interactions between the identified compounds and the target Mpro protein, molecular docking was performed using AutoDock 4.2. The findings identified compounds 4, 5, 10, and 14 as the most potential inhibitors of Mpro with binding energies of −9.3, −8.5, −8.1, and −7.8 kcal mol −1, respectively. In order to assess the stability of the protein–ligand complexes, molecular dynamics simulations were conducted for a duration of 100 ns, and various parameters such as RMSD, RMSF, Rg, and SASA were evaluated. All selected compounds 4, 5, 10, and 14 showed considerable Mpro inhibiting activity in vitro, with compound 4 being the most powerful with an IC 50 value of 1.2 µg/mL. MM-GBSA free energy calculations also revealed compound 4 as the most powerful Mpro inhibitor. None of the compounds ( 4, 5, 10, and 14) display any significant cytotoxic activity against A549 and HUVEC cell lines.
Related collections
Most cited references99
- Record: found
- Abstract: found
- Article: not found
A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19
- Record: found
- Abstract: found
- Article: not found
Structure of Mpro from COVID-19 virus and discovery of its inhibitors
- Record: found
- Abstract: found
- Article: found
