- Record: found
- Abstract: found
- Article: found
Insights into ultra-low affinity lipase-antibody noncovalent complex binding mechanisms
Read this article at
ABSTRACT
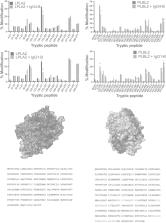
Detection of host cell protein (HCP) impurities is critical to ensuring that recombinant drug products, including monoclonal antibodies (mAbs), are safe. Mechanistic characterization as to how HCPs persist in drug products is important to refining downstream processing. It has been hypothesized that weak lipase–mAb interactions enable HCP lipases to evade drug purification processes. Here, we apply state-of-the-art methods to establish lipase-mAb binding mechanisms. First, the mass spectrometry (MS) approach of fast photochemical oxidation of proteins was used to elucidate putative binding regions. The CH1 domain was identified as a conserved interaction site for IgG1 and IgG4 mAbs against the HCPs phospholipase B-like protein (PLBL2) and lysosomal phospholipase A2 (LPLA2). Rationally designed mutations in the CH1 domain of the IgG4 mAb caused a 3- to 70-fold K D reduction against PLBL2 by surface plasmon resonance (SPR). LPLA2-IgG4 mutant complexes, undetected by SPR and studied using native MS collisional dissociation experiments, also showed significant complex disruption, from 16% to 100%. Native MS and ion mobility (IM) determined complex stoichiometries for four lipase-IgG4 complexes and directly interrogated the enrichment of specific lipase glycoforms. Confirmed with time-course and exoglycosidase experiments, deglycosylated lipases prevented binding, and low-molecular-weight glycoforms promoted binding, to mAbs. This work demonstrates the value of integrated biophysical approaches to characterize micromolar affinity complexes. It is the first in-depth structural report of lipase-mAb binding, finding roles for the CH1 domain and lipase glycosylation in mediating binding. The structural insights gained offer new approaches for the bioengineering of cells or mAbs to reduce HCP impurity levels.
Abbreviations: CAN, Acetonitrile; AMAC, Ammonium acetate; BFGS, Broyden–Fletcher–Goldfarb–Shanno; CHO, Chinese Hamster Ovary; K D, Dissociation constant; DTT, Dithiothreitol; ELISA, Enzyme-linked immunosorbent assay; FPOP, Fast photochemical oxidation of proteins; FA, Formic acid; F(ab’), Fragment antibodies; HCP, Host cell protein; IgG, Immunoglobulin; IM, Ion mobility; LOD, Lower limit of detection; LPLA2, Lysosomal phospholipase A2; Man, Mannose; MS, Mass spectrometry; MeOH, Methanol; MST, Microscale thermophoresis; mAbs, Monoclonal antibodies; PPT1, Palmitoyl protein thioesterase; ppm, Parts per million; PLBL2, Phospholipase B-like protein; PLD3, Phospholipase D3; PS-20, Polysorbate-20; SP, Sphingomyelin phosphodiesterase; SPR, Surface plasmon resonance; TFA, Trifluoroacetic acid.
Related collections
Most cited references60
- Record: found
- Abstract: found
- Article: not found
Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles.
- Record: found
- Abstract: not found
- Article: not found
MicroScale Thermophoresis: Interaction analysis and beyond

- Record: found
- Abstract: found
- Article: found