- Record: found
- Abstract: found
- Article: found
The interplay between electron transport chain function and iron regulatory factors influences melanin formation in Cryptococcus neoformans
Read this article at
Abstract
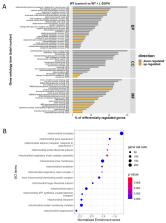
Mitochondrial functions are critical for the ability of the fungal pathogen Cryptococcus neoformans to cause disease. However, mechanistic connections between key functions such as the mitochondrial electron transport chain (ETC) and virulence factor elaboration have yet to be thoroughly characterized. Here, we observed that inhibition of ETC complex III suppressed melanin formation, a major virulence factor. This inhibition was partially blocked upon loss of Cir1 or HapX, two transcription factors that regulate iron acquisition and use. In this regard, loss of Cir1 derepresses the expression of laccase genes as a potential mechanism to restore melanin, while HapX may condition melanin formation by controlling oxidative stress. We hypothesize that ETC dysfunction alters redox homeostasis to influence melanin formation. Consistent with this idea, inhibition of growth by hydrogen peroxide was exacerbated in the presence of the melanin substrate L-DOPA. Additionally, loss of the mitochondrial chaperone Mrj1, which influences the activity of ETC complex III and reduces ROS accumulation, also partially blocked antimycin A inhibition of melanin. The phenotypic impact of mitochondrial dysfunction was consistent with RNA-Seq analyses of WT cells treated with antimycin A or L-DOPA, or cells lacking Cir1 that revealed influences on transcripts encoding mitochondrial functions (e.g., ETC components and proteins for Fe-S cluster assembly). Overall, these findings reveal mitochondria-nuclear communication via ROS and iron regulators to control virulence factor production in C. neoformans.
Related collections
Most cited references73

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles
- Record: found
- Abstract: found
- Article: found