- Record: found
- Abstract: found
- Article: found
Incorporating family history of disease improves polygenic risk scores in diverse populations

Read this article at
SUMMARY
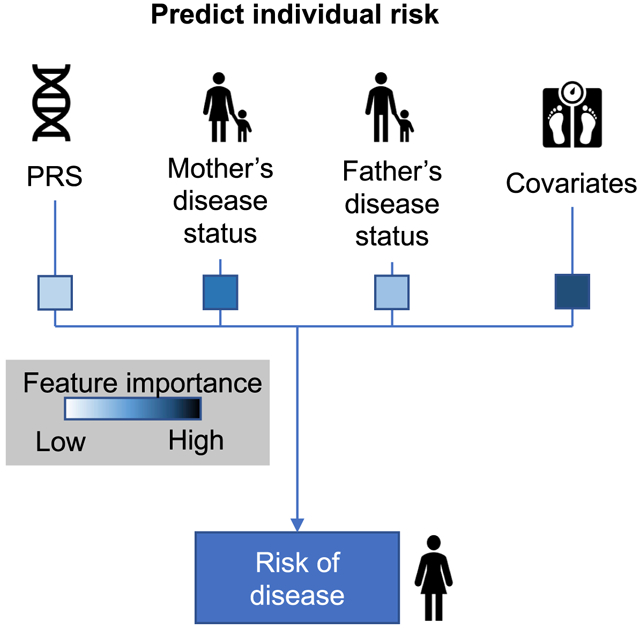
Polygenic risk scores (PRSs) derived from genotype data and family history (FH) of disease provide valuable information for predicting disease risk, but PRSs perform poorly when applied to diverse populations. Here, we explore methods for combining both types of information (PRS-FH) in UK Biobank data. PRSs were trained using all British individuals (n = 409,000), and target samples consisted of unrelated non-British Europeans (n = 42,000), South Asians (n = 7,000), or Africans (n = 7,000). We evaluated PRS, FH, and PRS-FH using liability-scale R 2, primarily focusing on 3 well-powered diseases (type 2 diabetes, hypertension, and depression). PRS attained average prediction R 2s of 5.8%, 4.0%, and 0.53% in non-British Europeans, South Asians, and Africans, confirming poor cross-population transferability. In contrast, PRS-FH attained average prediction R 2s of 13%, 12%, and 10%, respectively, representing a large improvement in Europeans and an extremely large improvement in Africans. In conclusion, including family history improves the accuracy of polygenic risk scores, particularly in diverse populations.
Graphical Abstract
In brief
Polygenic risk scores (PRSs) and family history (FH) of disease provide valuable information for predicting disease risk, but PRSs perform poorly when applied to diverse populations. Hujoel et al. explore methods for combining PRSs and FH and find that including FH improves prediction accuracy, particularly in diverse populations.
Related collections
Most cited references41

- Record: found
- Abstract: found
- Article: found
The UK Biobank resource with deep phenotyping and genomic data
- Record: found
- Abstract: found
- Article: not found
Clinical use of current polygenic risk scores may exacerbate health disparities
- Record: found
- Abstract: found
- Article: not found