- Record: found
- Abstract: found
- Article: found
Phosphorylation of Mcm2 modulates Mcm2–7 activity and affects the cell’s response to DNA damage
Read this article at
Abstract
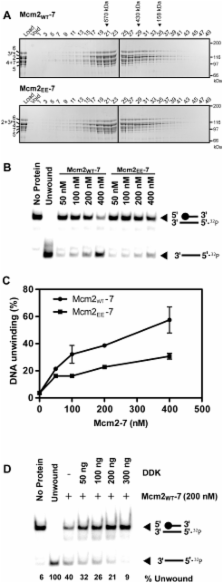
The S-phase kinase, DDK controls DNA replication through phosphorylation of the replicative helicase, Mcm2–7. We show that phosphorylation of Mcm2 at S164 and S170 is not essential for viability. However, the relevance of Mcm2 phosphorylation is demonstrated by the sensitivity of a strain containing alanine at these positions ( mcm2 AA ) to methyl methanesulfonate (MMS) and caffeine. Consistent with a role for Mcm2 phosphorylation in response to DNA damage, the mcm2 AA strain accumulates more RPA foci than wild type. An allele with the phosphomimetic mutations S164E and S170E ( mcm2 EE ) suppresses the MMS and caffeine sensitivity caused by deficiencies in DDK function. In vitro, phosphorylation of Mcm2 or Mcm2 EE reduces the helicase activity of Mcm2–7 while increasing DNA binding. The reduced helicase activity likely results from the increased DNA binding since relaxing DNA binding with salt restores helicase activity. The finding that the ATP site mutant mcm2 K549R has higher DNA binding and less ATPase than mcm2 EE , but like mcm2 AA results in drug sensitivity, supports a model whereby a specific range of Mcm2–7 activity is required in response to MMS and caffeine. We propose that phosphorylation of Mcm2 fine-tunes the activity of Mcm2–7, which in turn modulates DNA replication in response to DNA damage.
Related collections
Most cited references65
- Record: found
- Abstract: found
- Article: not found
Cell wall integrity signaling in Saccharomyces cerevisiae.
- Record: found
- Abstract: found
- Article: not found
New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites.
- Record: found
- Abstract: found
- Article: not found