- Record: found
- Abstract: found
- Article: found
The Emergence of Successful Streptococcus pyogenes Lineages through Convergent Pathways of Capsule Loss and Recombination Directing High Toxin Expression
Read this article at
Abstract
Streptococcus pyogenes is a genetically diverse pathogen, with over 200 different genotypes defined by emm typing, but only a minority of these genotypes are responsible for the majority of human infection in high-income countries. Two prevalent genotypes associated with disease rose to international dominance following recombination of a toxin locus that conferred increased expression. Here, we found that recombination of this locus and promoter has occurred in other diverse genotypes, events that may allow these genotypes to expand in the population. We identified an association between the loss of hyaluronic acid capsule synthesis and high toxin expression, which we propose may be associated with an adaptive advantage. As S. pyogenes pathogenesis depends both on capsule and toxin production, new variants with altered expression may result in abrupt changes in the molecular epidemiology of this pathogen in the human population over time.
ABSTRACT
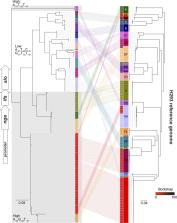
Gene transfer and homologous recombination in Streptococcus pyogenes has the potential to trigger the emergence of pandemic lineages, as exemplified by lineages of emm1 and emm89 that emerged in the 1980s and 2000s, respectively. Although near-identical replacement gene transfer events in the nga (NADase) and slo (streptolysin O) loci conferring high expression of these toxins underpinned the success of these lineages, extension to other emm genotype lineages is unreported. The emergent emm89 lineage was characterized by five regions of homologous recombination additional to nga-slo, including complete loss of the hyaluronic acid capsule synthesis locus hasABC, a genetic trait replicated in two other leading emm types and recapitulated by other emm types by inactivating mutations. We hypothesized that other leading genotypes may have undergone similar recombination events. We analyzed a longitudinal data set of genomes from 344 clinical invasive disease isolates representative of locations across England, dating from 2001 to 2011, and an international collection of S. pyogenes genomes representing 54 different genotypes and found frequent evidence of recombination events at the nga- slo locus predicted to confer higher toxin genotype. We identified multiple associations between recombination at this locus and inactivating mutations within hasAB, suggesting convergent evolutionary pathways in successful genotypes. This included common genotypes emm28 and emm87. The combination of no or low capsule and high expression of nga and slo may underpin the success of many emergent S. pyogenes lineages of different genotypes, triggering new pandemics, and could change the way S. pyogenes causes disease.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Global emm type distribution of group A streptococci: systematic review and implications for vaccine development.

- Record: found
- Abstract: found
- Article: found
Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data
- Record: found
- Abstract: not found
- Article: not found