- Record: found
- Abstract: found
- Article: found
Integrating cryo-OrbiSIMS with computational modelling and metadynamics simulations enhances RNA structure prediction at atomic resolution
Read this article at
Abstract
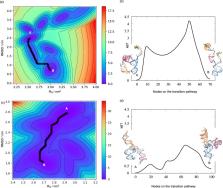
The 3D architecture of RNAs governs their molecular interactions, chemical reactions, and biological functions. However, a large number of RNAs and their protein complexes remain poorly understood due to the limitations of conventional structural biology techniques in deciphering their complex structures and dynamic interactions. To address this limitation, we have benchmarked an integrated approach that combines cryogenic OrbiSIMS, a state-of-the-art solid-state mass spectrometry technique, with computational methods for modelling RNA structures at atomic resolution with enhanced precision. Furthermore, using 7SK RNP as a test case, we have successfully determined the full 3D structure of a native RNA in its apo, native and disease-remodelled states, which offers insights into the structural interactions and plasticity of the 7SK complex within these states. Overall, our study establishes cryo-OrbiSIMS as a valuable tool in the field of RNA structural biology as it enables the study of challenging, native RNA systems.
Abstract
Conventional structural biology techniques are limited in deciphering complex RNA structures and dynamic interactions. Here the authors show an integrated approach that combines cryogenic OrbiSIMS (cryo-OrbiSIMS) with computational methods for modelling RNA structures at atomic resolution.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.
- Record: found
- Abstract: not found
- Article: not found
GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers

- Record: found
- Abstract: found
- Article: found