- Record: found
- Abstract: found
- Article: not found
DNA Methylation in the Human Cerebral Cortex Is Dynamically Regulated throughout the Life Span and Involves Differentiated Neurons
Read this article at
Abstract
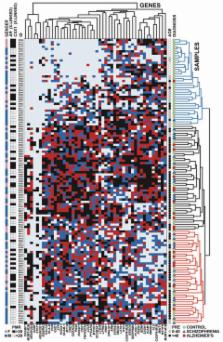
The role of DNA cytosine methylation, an epigenetic regulator of chromatin structure and function, during normal and pathological brain development and aging remains unclear. Here, we examined by MethyLight PCR the DNA methylation status at 50 loci, encompassing primarily 5′ CpG islands of genes related to CNS growth and development, in temporal neocortex of 125 subjects ranging in age from 17 weeks of gestation to 104 years old. Two psychiatric disease cohorts—defined by chronic neurodegeneration (Alzheimer's) or lack thereof (schizophrenia)—were included. A robust and progressive rise in DNA methylation levels across the lifespan was observed for 8/50 loci ( GABRA2, GAD1, HOXA1, NEUROD1, NEUROD2, PGR, STK11, SYK) typically in conjunction with declining levels of the corresponding mRNAs. Another 16 loci were defined by a sharp rise in DNA methylation levels within the first few months or years after birth. Disease-associated changes were limited to 2/50 loci in the Alzheimer's cohort, which appeared to reflect an acceleration of the age-related change in normal brain. Additionally, methylation studies on sorted nuclei provided evidence for bidirectional methylation events in cortical neurons during the transition from childhood to advanced age, as reflected by significant increases at 3, and a decrease at 1 of 10 loci. Furthermore, the DNMT3a de novo DNA methyl-transferase was expressed across all ages, including a subset of neurons residing in layers III and V of the mature cortex. Therefore, DNA methylation is dynamically regulated in the human cerebral cortex throughout the lifespan, involves differentiated neurons, and affects a substantial portion of genes predominantly by an age-related increase.
Related collections
Most cited references43
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found
Epigenetic programming by maternal behavior.
- Record: found
- Abstract: found
- Article: not found
