- Record: found
- Abstract: found
- Article: found
Comparative paleovirological analysis of crustaceans identifies multiple widespread viral groups
Read this article at
Abstract
Background
The discovery of many fragments of viral genomes integrated in the genome of their eukaryotic host (endogenous viral elements; EVEs) has recently opened new avenues to further our understanding of viral evolution and of host-virus interactions. Here, we report the results of a comprehensive screen for EVEs in crustaceans. Following up on the recent discovery of EVEs in the terrestrial isopod, Armadillidium vulgare, we scanned the genomes of six crustacean species: a terrestrial isopod ( Armadillidium nasatum), two water fleas ( Daphnia pulex and D. pulicaria), two copepods (the salmon louse, Lepeophtheirus salmonis and Eurytemora affinis), and a freshwater amphipod ( Hyalella azteca).
Results
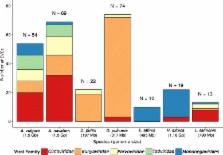
In total, we found 210 EVEs representing 14 different lineages belonging to five different viral groups that are present in two to five species: Bunyaviridae (−ssRNA), Circoviridae (ssDNA), Mononegavirales (−ssRNA), Parvoviridae (ssDNA) and Totiviridae (dsRNA). The identification of shared orthologous insertions between A. nasatum and A. vulgare indicates that EVEs have been maintained over several millions of years, although we did not find any evidence supporting exaptation. Overall, the different degrees of EVE degradation (from none to >10 nonsense mutations) suggest that endogenization has been recurrent during the evolution of the various crustacean taxa. Our study is the first to report EVEs in D. pulicaria, E. affinis and H. azteca, many of which are likely to result from recent endogenization of currently circulating viruses.
Conclusions
In conclusion, we have unearthed a large diversity of EVEs from crustacean genomes, and shown that four of the five viral groups we uncovered ( Bunyaviridae, Circoviridae, Mononegavirales, Parvoviridae) were and may still be present in three to four highly divergent crustacean taxa. In addition, the discovery of recent EVEs offers an interesting opportunity to characterize new exogenous viruses currently circulating in economically or ecologically important copepod species.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers.
- Record: found
- Abstract: not found
- Article: not found
Le piégeage lumineux, moyen d'approche de la faune entomologique d'un grand fleuve (Ephéméroptères, en particulier)
- Record: found
- Abstract: found
- Article: not found