- Record: found
- Abstract: found
- Article: found
Emergence of an erythroid cell-specific regulatory region in ABO intron 1 attributable to A- or B-antigen expression on erythrocytes in Hominoidea
Read this article at
Abstract
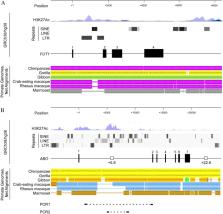
A- and B-antigens are present on red blood cells (RBCs) as well as other cells and secretions in Hominoidea including humans and apes such as chimpanzees and gibbons, whereas expression of these antigens on RBCs is subtle in monkeys such as Japanese macaques. Previous studies have indicated that H-antigen expression has not completely developed on RBCs in monkeys. Such antigen expression requires the presence of H-antigen and A- or B-transferase expression in cells of erythroid lineage, although whether or not ABO gene regulation is associated with the difference of A- or B-antigen expression between Hominoidea and monkeys has not been examined. Since it has been suggested that ABO expression on human erythrocytes is dependent upon an erythroid cell-specific regulatory region or the + 5.8-kb site in intron 1, we compared the sequences of ABO intron 1 among non-human primates, and demonstrated the presence of sites orthologous to the + 5.8-kb site in chimpanzees and gibbons, and their absence in Japanese macaques. In addition, luciferase assays revealed that the former orthologues enhanced promoter activity, whereas the corresponding site in the latter did not. These results suggested that the A- or B-antigens on RBCs might be ascribed to emergence of the + 5.8-kb site or the corresponding regions in ABO through genetic evolution.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Molecular genetic basis of the histo-blood group ABO system.

- Record: found
- Abstract: found
- Article: found
Classification and characterization of human endogenous retroviruses; mosaic forms are common
- Record: found
- Abstract: found
- Article: not found