- Record: found
- Abstract: found
- Article: not found
The long-range interaction landscape of gene promoters
Read this article at
Abstract
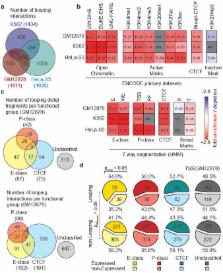
The vast non-coding portion of the human genome is awash in functional elements and disease-causing regulatory variants. The principles defining the relationships between these elements and distal target genes remain unknown. Promoters and distal elements can engage in looping interactions that have been implicated in gene regulation 1 . Here we have applied chromosome conformation capture carbon copy, 5C 2 , to comprehensively interrogate interactions between transcription start sites (TSSs) and distal elements in 1% of the human genome representing the ENCODE pilot project regions 3 . 5C maps were generated for GM12878, K562 and HeLa-S3 cells and results were integrated with data from the ENCODE consortium 4 . In each cell line we discovered >1,000 long-range interactions between promoters and distal sites that include elements resembling enhancers, promoters and CTCF-bound sites. We observed significant correlations between gene expression, promoter-enhancer interactions and the presence of enhancer RNAs. Long-range interactions display striking asymmetry with a bias for interactions with elements located ~120 Kb upstream of the TSS. Long-range interactions are often not blocked by sites bound by CTCF and cohesin implying that many of these sites do not demarcate physically insulated gene domains. Further, only ~7% of looping interactions are with the nearest gene, suggesting that genomic proximity is not a simple predictor for long-range interactions. Finally, promoters and distal elements are engaged in multiple long-range interactions to form complex networks. Our results start to place genes and regulatory elements in three-dimensional context, revealing their functional relationships.
Related collections
Most cited references21
- Record: found
- Abstract: found
- Article: not found
Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements.
- Record: found
- Abstract: found
- Article: not found
Cohesin mediates transcriptional insulation by CCCTC-binding factor.

- Record: found
- Abstract: found
- Article: found
