- Record: found
- Abstract: found
- Article: found
A neutral invertase controls cell division besides hydrolysis of sucrose for nutrition during germination and seed setting in rice

Read this article at
Summary
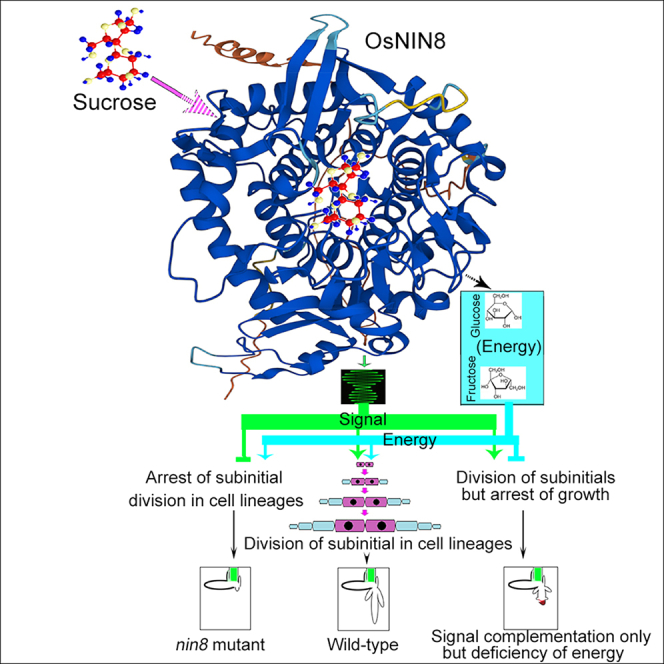
Sucrose is the transport form of carbohydrate in plants serving as signal molecule besides nutrition, but the signaling is elusive. Here, neutral invertase 8 (OsNIN8) mutated at G461R into OsNIN8m, which increased its charge and hydrophobicity, decreased hydrolysis of sucrose to 13% and firmer binding to sucrose than the wildtype. This caused downstream metabolites and energy accumulation forming overnutrition. Paradoxically, division of subinitials in longitudinal cell lineages was only about 15 times but more than 100 times in wildtype, resulting in short radicle. Further, mutation of OsNIN8 into deficiency of hydrolysis but maintenance of sucrose binding allowed cell division until ran out of energy showing the association but not hydrolysis gave the signal. Chemically, sucrose binding to OsNIN8 was exothermic but to OsNIN8m was endothermic. Therefore, OsNIN8m lost the signal function owing to change of thermodynamic state. So, OsNIN8 sensed sucrose for cell division besides hydrolyzed sucrose.
Graphical abstract
Highlights
-
•
OsNIN8 binds to and hydrolyzes sucrose for cell division in root tip during germination
-
•
OsNIN8 Wildtype exothermic association with sucrose initiates signal for cell division
-
•
The signal controls cell division of subinitials in longitudinal cell lineages
-
•
Sucrose roles of nutrition and signal can be separated in different mutants
Abstract
Cell biology; Plant biology; Plant development
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
Sucrose metabolism: gateway to diverse carbon use and sugar signaling.
- Record: found
- Abstract: found
- Article: not found
Genetic compensation triggered by mutant mRNA degradation
- Record: found
- Abstract: not found
- Article: not found