- Record: found
- Abstract: found
- Article: not found
A Specificity Map for the PDZ Domain Family
Read this article at
Abstract
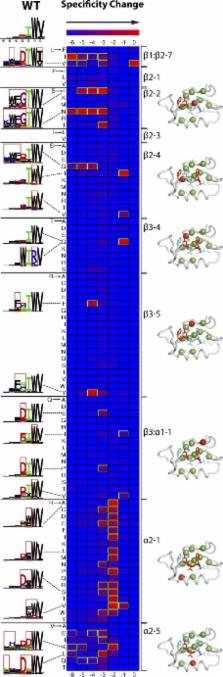
PDZ domains are protein–protein interaction modules that recognize specific C-terminal sequences to assemble protein complexes in multicellular organisms. By scanning billions of random peptides, we accurately map binding specificity for approximately half of the over 330 PDZ domains in the human and Caenorhabditis elegans proteomes. The domains recognize features of the last seven ligand positions, and we find 16 distinct specificity classes conserved from worm to human, significantly extending the canonical two-class system based on position −2. Thus, most PDZ domains are not promiscuous, but rather are fine-tuned for specific interactions. Specificity profiling of 91 point mutants of a model PDZ domain reveals that the binding site is highly robust, as all mutants were able to recognize C-terminal peptides. However, many mutations altered specificity for ligand positions both close and far from the mutated position, suggesting that binding specificity can evolve rapidly under mutational pressure. Our specificity map enables the prediction and prioritization of natural protein interactions, which can be used to guide PDZ domain cell biology experiments. Using this approach, we predicted and validated several viral ligands for the PDZ domains of the SCRIB polarity protein. These findings indicate that many viruses produce PDZ ligands that disrupt host protein complexes for their own benefit, and that highly pathogenic strains target PDZ domains involved in cell polarity and growth.
Author Summary
The PDZ domain is a structural domain that functions as a protein–protein interaction module that recognizes specific C-terminal peptide sequences to assemble intracellular complexes important in signaling pathways of multicellular organisms. These modules are associated with human disease and are targets of viruses and other pathogens. By examining peptide specificity and substrate diversity of roughly one half of the PDZ domains known to exist in human and the nematode Caenorhabditis elegans, we were able to show that PDZ domains are more specific than previously appreciated. PDZ domains also remain functional under high mutational pressure, and only a few of the vast number of possible PDZ domain specificities are utilized in nature. These PDZ domain specificities are conserved from human to worm, implying that the specificities evolved early and were reused over evolution instead of being reshaped. The specificity map generated here was used to predict and experimentally confirm new viral PDZ-binding motifs. We present evidence that pathogenic viruses, including avian influenza, bind host PDZ domains via these motifs, thereby competing with signaling by host complexes, which leads to disruption of growth and polarity of the host cells.
Abstract
A genome-scale specificity map for PDZ domains reveals how family members recognize ligands to assemble signaling complexes and also reveals how viruses target these domains to subvert host cell function.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Ensembl 2007
- Record: found
- Abstract: found
- Article: not found
