- Record: found
- Abstract: found
- Article: found
The N-terminal Helix-Turn-Helix Motif of Transcription Factors MarA and Rob Drives DNA Recognition
Read this article at
Abstract

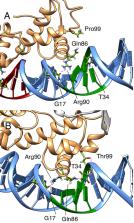
DNA-binding proteins play an important role in gene regulation and cellular function. The transcription factors MarA and Rob are two homologous members of the AraC/XylS family that regulate multidrug resistance. They share a common DNA-binding domain, and Rob possesses an additional C-terminal domain that permits binding of low-molecular weight effectors. Both proteins possess two helix-turn-helix (HTH) motifs capable of binding DNA; however, while MarA interacts with its promoter through both HTH-motifs, prior studies indicate that Rob binding to DNA via a single HTH-motif is sufficient for tight binding. In the present work, we perform microsecond time scale all-atom simulations of the binding of both transcription factors to different DNA sequences to understand the determinants of DNA recognition and binding. Our simulations characterize sequence-dependent changes in dynamical behavior upon DNA binding, showcasing the role of Arg40 of the N-terminal HTH-motif in allowing for specific tight binding. Finally, our simulations demonstrate that an acidic C-terminal loop of Rob can control the DNA binding mode, facilitating interconversion between the distinct DNA binding modes observed in MarA and Rob. In doing so, we provide detailed molecular insight into DNA binding and recognition by these proteins, which in turn is an important step toward the efficient design of antivirulence agents that target these proteins.
Related collections
Most cited references95
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.
- Record: found
- Abstract: not found
- Article: not found
VMD: Visual molecular dynamics
- Record: found
- Abstract: found
- Article: not found