- Record: found
- Abstract: found
- Article: not found
Poly (ADP-Ribose) Polymerase 1 Is Required for Protein Localization to Cajal Body
Read this article at
Abstract
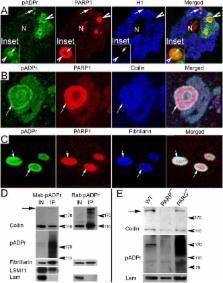
Recently, the nuclear protein known as Poly (ADP-ribose) Polymerase1 (PARP1) was shown to play a key role in regulating transcription of a number of genes and controlling the nuclear sub-organelle nucleolus. PARP1 enzyme is known to catalyze the transfer of ADP-ribose to a variety of nuclear proteins. At present, however, while we do know that the main acceptor for pADPr in vivo is PARP1 protein itself, by PARP1 automodification, the significance of PARP1 automodification for in vivo processes is not clear. Therefore, we investigated the roles of PARP1 auto ADP-ribosylation in dynamic nuclear processes during development. Specifically, we discovered that PARP1 automodification is required for shuttling key proteins into Cajal body (CB) by protein non-covalent interaction with pADPr in vivo. We hypothesize that PARP1 protein shuttling follows a chain of events whereby, first, most unmodified PARP1 protein molecules bind to chromatin and accumulate in nucleoli, but then, second, upon automodification with poly(ADP-ribose), PARP1 interacts non-covalently with a number of nuclear proteins such that the resulting protein-pADPr complex dissociates from chromatin into CB.
Author Summary
Previous studies revealed vital roles for the Poly(ADP-ribose) Polymerase (PARP) protein in developmental program regulation, as well as in chromatin loosening and transcriptional activation. The main gap in our understanding of PARP protein roles during development involves integrating the biological activities of PARP protein into a general network of nuclear protein dynamics and gene expression. Here, we report the functional association of PARP protein activity with proteins trafficking through Cajal bodies. Cajal bodies (CBs) are nuclear organelles that regulate the biogenesis of RNA-protein complexes involved in transcription and splicing. We demonstrate that (1) PARP is present in CBs, (2) its loss leads to CB disassembly, (3) an increase in PARP levels leads to formation of CBs, and, finally, (4) PARP localization is ecdysone sensitive. We further propose a model in which PARP controls protein trafficking through the CB and contributes to CB formation.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions.
- Record: found
- Abstract: found
- Article: not found
Transposition of cloned P elements into Drosophila germ line chromosomes.
- Record: found
- Abstract: found
- Article: not found
