- Record: found
- Abstract: found
- Article: found
The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
Read this article at
Abstract
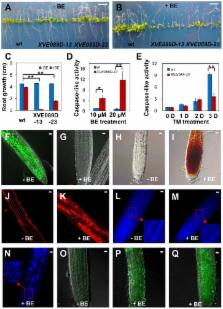
The unfolded protein response (UPR) is activated to sustain cell survival by reducing misfolded protein accumulation in the endoplasmic reticulum (ER). The UPR also promotes programmed cell death (PCD) when the ER stress is severe; however, the underlying molecular mechanisms are less understood, especially in plants. Previously, two membrane-associated transcriptions factors (MTFs), bZIP28 and bZIP60, were identified as the key regulators for cell survival in the plant ER stress response. Here, we report the identification of another MTF, NAC089, as an important PCD regulator in Arabidopsis ( Arabidopsis thaliana) plants. NAC089 relocates from the ER membrane to the nucleus under ER stress conditions. Inducible expression of a truncated form of NAC089, in which the transmembrane domain is deleted, induces PCD with increased caspase 3/7-like activity and DNA fragmentation. Knock-down NAC089 in Arabidopsis confers ER stress tolerance and impairs ER-stress-induced caspase-like activity. Transcriptional regulation analysis and ChIP-qPCR reveal that NAC089 plays important role in regulating downstream genes involved in PCD, such as NAC094, MC5 and BAG6. Furthermore, NAC089 is up-regulated by ER stress, which is directly controlled by bZIP28 and bZIP60. These results show that nuclear relocation of NAC089 promotes ER-stress-induced PCD, and both pro-survival and pro-death signals are elicited by bZIP28 and bZIP60 during plant ER stress response.
Author Summary
Protein folding is fundamentally important for development and responses to environmental stresses in eukaryotes. When excess misfolded proteins are accumulated in the endoplasmic reticulum (ER), the unfolded protein response (UPR) is triggered to promote cell survival through optimizing protein folding, and also promote programmed cell death (PCD) when the stress is severe. However, the link from ER-stress-sensing to PCD is largely unknown. Here, we report the identification of one membrane-associated transcription factor NAC089 as an important regulator of ER stress-induced PCD in plants. We have established a previously unrecognized molecular connection between ER stress sensors and PCD regulators. We have shown that organelle-to-organelle translocation of a transcription factor is important for its function in transcriptional regulation. Our results have provided novel insights into the molecular mechanisms of PCD in plants, especially under ER stress conditions.
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants
- Record: found
- Abstract: found
- Article: not found
Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase.
- Record: found
- Abstract: found
- Article: not found