- Record: found
- Abstract: found
- Article: found
Bidirectional interplay between SARS-CoV-2 and autophagy
Read this article at
ABSTRACT
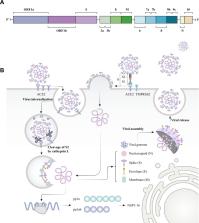
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), as the causative agent of the recent COVID-19 pandemic, continues representing one of the main health concerns worldwide. Autophagy, in addition to its role in cellular homeostasis and metabolism, plays an important part for the host antiviral immunity. However, viruses including SARS-CoV-2 have evolved diverse mechanisms to not only overcome autophagy’s antiviral pressure but also manipulate its machinery in order to enhance viral replication and propagation. Here, we discuss our current knowledge on the impact that autophagy exerts on SARS-CoV-2 replication, as well as the different counteracting measures that this virus has developed to manipulate autophagy’s complex machinery. Some of the elements regarding this interplay may become future therapeutic targets in the fight against SARS-CoV-2.
Related collections
Most cited references112
- Record: found
- Abstract: found
- Article: not found
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor
- Record: found
- Abstract: found
- Article: not found
