- Record: found
- Abstract: found
- Article: found
Epigenetics of Male Infertility: The Role of DNA Methylation
Read this article at
Abstract
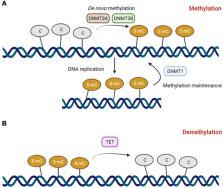
In recent years, a number of studies focused on the role of epigenetics, including DNA methylation, in spermatogenesis and male infertility. We aimed to provide an overview of the knowledge concerning the gene and genome methylation and its regulation during spermatogenesis, specifically in the context of male infertility etiopathogenesis. Overall, the findings support the hypothesis that sperm DNA methylation is associated with sperm alterations and infertility. Several genes have been found to be differentially methylated in relation to impaired spermatogenesis and/or reproductive dysfunction. Particularly, DNA methylation defects of MEST and H19 within imprinted genes and MTHFR within non-imprinted genes have been repeatedly linked with male infertility. A deep knowledge of sperm DNA methylation status in association with reduced reproductive potential could improve the development of novel diagnostic tools for this disease. Further studies are needed to better elucidate the mechanisms affecting methylation in sperm and their impact on male infertility.
Related collections
Most cited references235
- Record: found
- Abstract: found
- Article: not found
DNA methylation and its basic function.
- Record: found
- Abstract: found
- Article: not found
The diverse roles of DNA methylation in mammalian development and disease
- Record: found
- Abstract: found
- Article: not found