- Record: found
- Abstract: found
- Article: found
miR-15b reduces amyloid-β accumulation in SH-SY5Y cell line through targetting NF-κB signaling and BACE1
Read this article at
Abstract
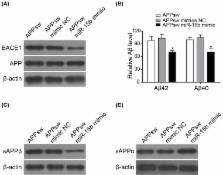
Alzheimer’s disease (AD) is the multifactorial neurodegenerative disorder causing progressive memory loss and cognitive impairment. The aberrant accumulation of amyloid-β (Aβ) and neuroinflammation are two major events in AD. BACE1 is required for the cleavage of amyloid precursor protein (APP) to generate Aβ, which stimulates the nuclear transcription factor κB (NF-κB) signaling, leading to the secretion of inflammatory cytokines. And NF-κB can up-regulate the expression of BACE1. miRNAs are small non-coding RNAs that regulate gene transcription. miR-15b down-regulates BACE1 expression while it is unclear whether miR-15b can regulate Aβ in human neuronal cells, and if so, whether it is by targetting NF-κB. SH-SY5Y cell line was transfected with Swedish APP mutant (APPswe) as an in vitro AD model. Quantitative PCR (qPCR), WB, and ELISA were used to detected related gene expression intracellularly or in supernatant. Dual luciferase assay was used to validate miRNA and targets binding. miR-15b inhibits expression of BACE1and APP. Moreover, the reduced level of Aβ was observed in response to miR-15b mimics in SH-SH5Y/APPswe cells. miR-15b directly targetted the conserved Bace1 3′UTR to regulate its expression. In addition, the inhibition of APPswe-induced secretion of inflammatory cytokines and the suppression of NF-κB activation by miR-15b were validated. And miR-15b directly targetted the 3′UTRs of NF-κB1 and inhibitor of NF-κB (IκB) kinase α (IKK-α), encoding NF-κB1 and IKK-α, respectively. Our study suggests that miR-15b inhibits Aβ accumulation through targetting NF-κB signaling and BACE1 and serves as a potential molecular target for AD therapy.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells.
- Record: found
- Abstract: found
- Article: not found
The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases.
